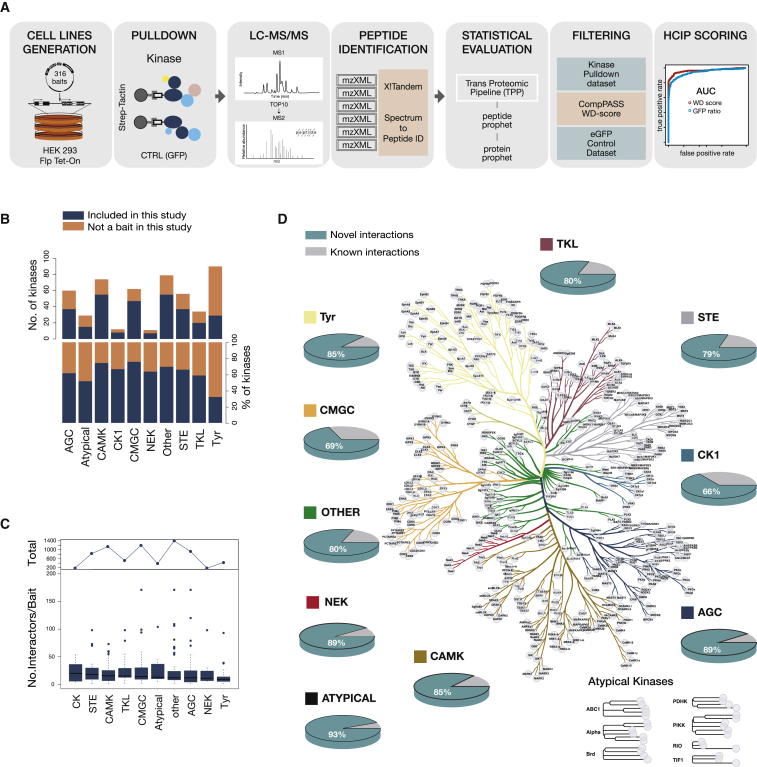
Figure 1.
Systematic Mapping of Kinase Interactions by AP-MS
(A) Workflow used to generate the kinase interaction network. This includes generation of more than 300 cell lines stably expressing doxycycline-inducible kinases, single-step pull-downs, duplicate runs on a hybrid linear ion trap-Orbitrap mass spectrometer, peptide identification with X!Tandem, and statistical evaluation.
(B) Coverage of kinases across different families. Dark blue represents included and orange not included kinases.
(C) Distribution of the number of identified interactors per kinase.
(D) Novel versus known interactions for each kinase family are plotted against the kinome evolutionary tree. Each kinase family is represented with a different color, and the same coloring scheme is used in all figures.