Figure 3.
Functional Landscape of the Kinase Interaction Network
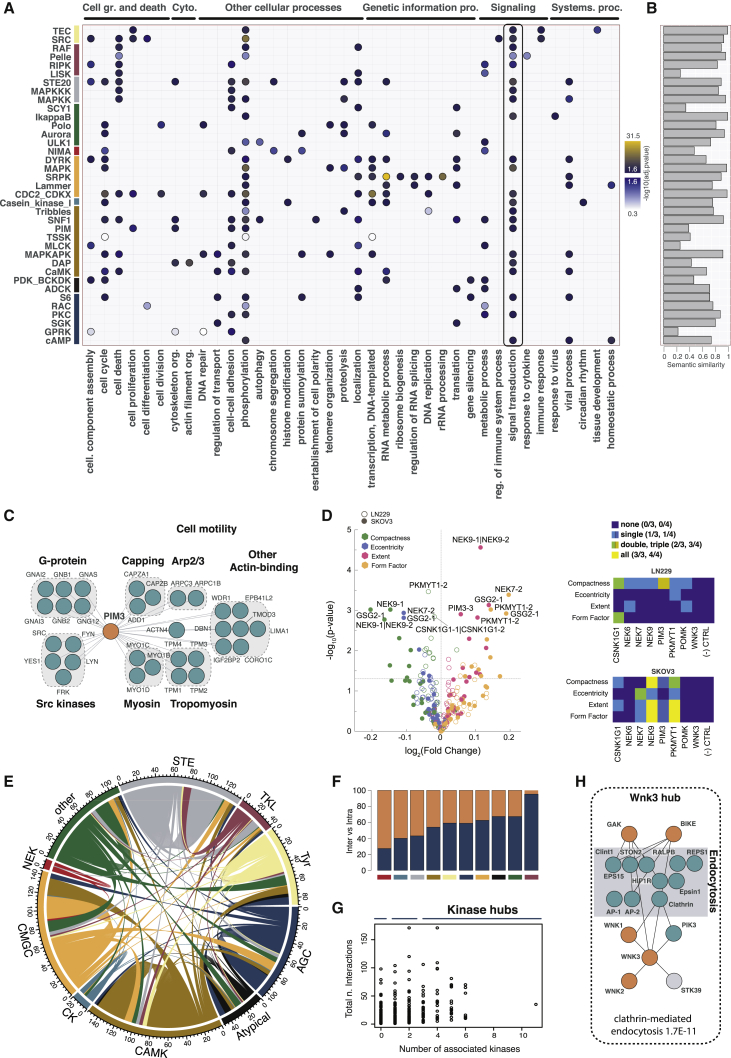
(A) Dotplot of the most significant terms associated with kinase subgroups covered in this study.
(B) Semantic similarity between the top GO terms based on the interactions from this study and interaction partners deposited in the IID.
(C) Main associations in the PIM3 interaction network include the actin cytoskeletal and G proteins and SRC modules.
(D) Cellular phenotyping after gene silencing. Volcano plot depicts siRNA gene targets that displayed the strongest changes in the representative cell area and shape phenotypes relative to the negative siRNA control. Targets that had a −log10 (p value) greater than 2.5 are labeled with their gene names. Colors represent different features measured, as defined by the CellProfiler software tool. Different siRNAs for the same gene are indicated with −1 or −2, and combinations of siRNAs are denoted by the symbol “|”. Heatmaps summarize the information on how many siRNA and siRNA combinations per gene were observed as significant.
(E) Circos plot of all kinase-kinase interactions. Coloring scheme is the same as in Figure 1.
(F) Ratio between intra-family and inter-family connections.
(G) Number of kinases versus total number of interactors per kinase bait.
(H) WNK3 kinase hub with its interactors and known associations with the endocytosis proteins.