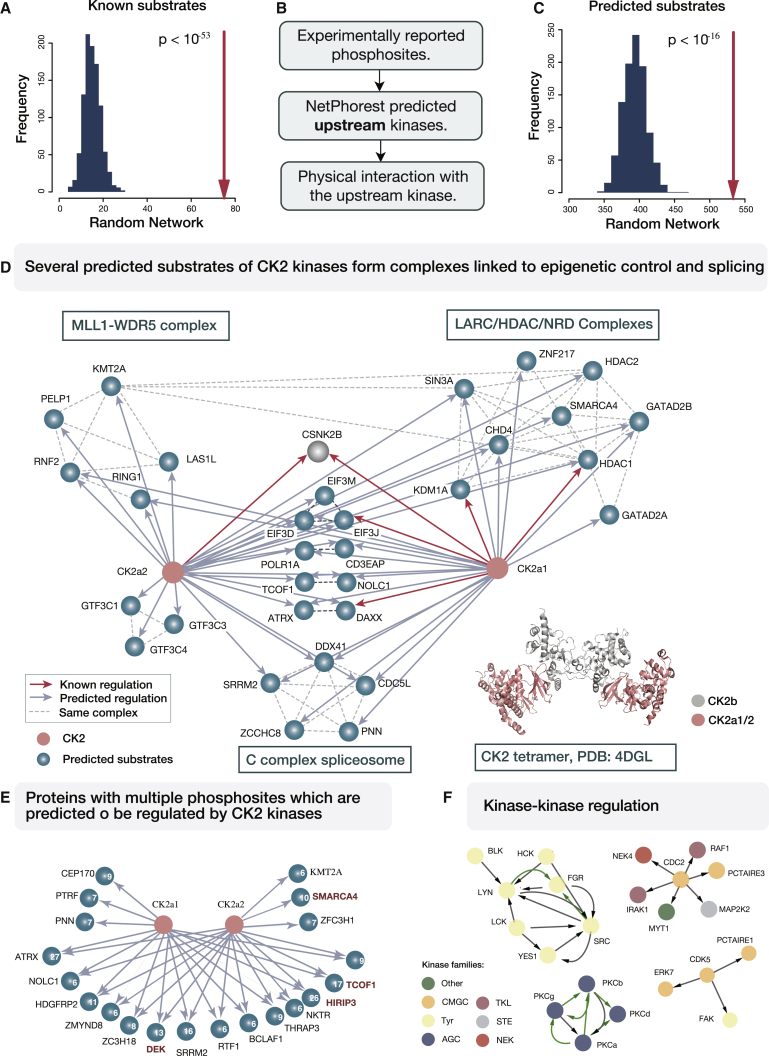
Figure 4.
Kinase Interaction Network Assists Assignments of Regulatory Interactions
(A) In the generated kinase network, 75 interactions (red arrow) were known kinase-substrate pairs, which corresponds to a significant enrichment compared to random networks (shown as a histogram on the left).
(B) Criteria for the prediction of novel kinase-substrate relationships.
(C) In total, 534 interactions in the kinase network were predicted as possible kinase-substrate interaction (depicted with red arrow). This is higher than the average number of predicted pairs in random networks (histogram on the left).
(D) Several substrate proteins predicted to be regulated by casein kinase 2 form stable protein complexes. Known and predicted regulatory interactions are depicted with red and gray arrows, respectively. Dashed lines indicate connections between proteins from the same CORUM complex (Ruepp et al., 2010). CK2 holoenzyme is a tetramer with two CK2b regulatory subunits and CK2a1 and CK2a2 catalytically active subunits.
(E) CK2 predicted substrates with more than five CK2 phosphomotifs are shown. Number of predicted phosphomotifs is shown for each protein. Proteins that were reported as CK2 substrates in previous studies are indicated in dark red.
(F) Kinase-kinase regulatory interactions occur within and across kinase families. The full set of predicted regulatory events is shown in Figure S3B. Arrows colored in green represent activation loop phosphorylation. Kinase families are colored according to the scheme in Figure 1.