Figure S2.
A Conserved Residency Profile for CD4 T Cells and Regulatory T Cells in the Healthy Mouse and Human Brain, Related to Figure 2
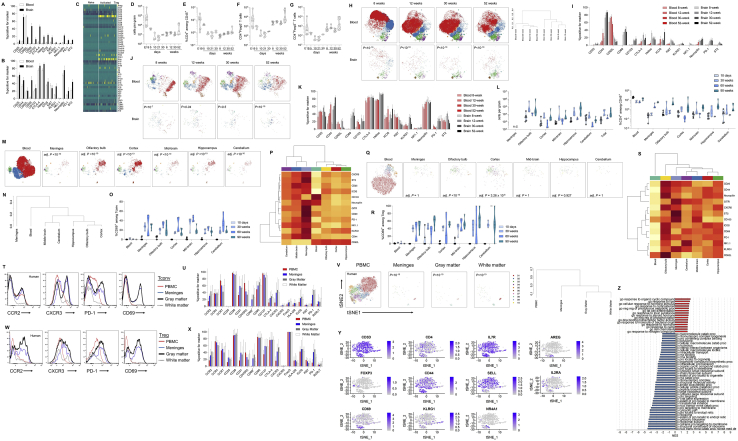
(A) Healthy perfused mouse brains were compared to blood by high-dimensional flow cytometry. Comparison of expression levels of CD62L, CD44, CD103, CD69, CD25, PD-1, Nrp1, ICOS, KLRG1, ST2, Ki67, Helios, T-bet and CTLA4 on blood versus brain CD4+CD3+CD45+CD8-Foxp3- T cells or (B) CD4+CD3+CD45+CD8-Foxp3+ Tregs (n = 5). (C) Transcription profile of CD4+ T cells purified from the murine brain, with analysis through the 10X single cell pipeline and filtering for known cytokines. Naive, activated and regulatory cells were defined based on tSNE clusters and the relative expression of CD44, CD62L and Foxp3 within each cluster. (D) CD4 T cells were assessed in the perfused mouse brain by high-dimensional flow cytometry. Wild-type mice were sampled across the late embryonic (day 19), post-natal development (day 5, 10, 21, 30) and during healthy aging (weeks 8, 12, 30 and 52). n = 9,3,3,5,2,8,5,6,5. Quantification of CD4+ cells per gram of brain tissue, (E) CD4+ T cells, as percentage of CD45+ cells, (F) CD4+Foxp3- conventional T cells and (G) CD4+Foxp3+ regulatory T cells. (H) tSNE of CD4+Foxp3- T cells gated on CD4+Foxp3-CD3+CD8-CD45+ cells and built on CD62L, CD44, CD103, CD69, CD25, PD-1, Nrp1, ICOS, KLRG1, ST2, Ki67, Helios, T-bet and CTLA4. FlowSOM clusters identified in color. P values refer to cross-entropy difference between age-matched blood and brain samples. Dendrogram represents cross-entropy distance between samples. (I) Comparison of expression levels of CD62L, CD44, CD103, CD69, CD25, PD-1, Nrp1, ICOS, KLRG1, ST2, Ki67, Helios, T-bet and CTLA4 on blood versus brain CD4+CD3+CD45+CD8-Foxp3- T cells at different ages. (G) tSNE of CD4+Foxp3+ T cells gated on CD4+Foxp3+CD3+CD8-CD45+ cells and built on CD62L, CD44, CD103, CD69, CD25, PD-1, Nrp1, ICOS, KLRG1, ST2, Ki67, Helios, T-bet and CTLA4. FlowSOM clusters identified in color. P values refer to cross-entropy difference between age-matched blood and brain samples. (J) Comparison of expression levels of CD62L, CD44, CD103, CD69, CD25, PD-1, Nrp1, ICOS, KLRG1, ST2, Ki67, Helios, T-bet and CTLA4 on blood versus brain CD4+CD3+CD45+CD8-Foxp3+ T cells at different ages. (K) Brain regions were surgically dissected and resident CD4 T cells were characterized by high-dimensional flow cytometry at 10 days, 30 weeks, 60 weeks and 90 weeks of age (n = 6,4,4,5). (L) Numbers and frequencies of CD4 T cells across brain regions in pups and adult mice. (M) tSNE of CD4+ Foxp3- T cells gated on CD4+Foxp3-CD3+CD8-CD45+ cells and built on CD62L, CD44, CD103, CD69, CD25, PD-1, Nrp1, ICOS, KLRG1, ST2, Ki67, Helios, T-bet, CTLA4, shown for blood and different brain regions in adult mice. The adjusted P value reflects the cross-entropy difference between tSNE plots in brain region versus blood. (N) Dendrogram showing the relationship in Tconv across the brain regions based on cross-entropy differences in tSNE. (O) CD69 expression in Tconv from the brain regions in pups and adult mice. (P) Heatmap showing expression of markers in brain region Tconv. (Q) tSNE of CD4+ Foxp3+ T cells gated on CD4+Foxp3+CD3+CD8-CD45+ cells and built on CD62L, CD44, CD103, CD69, CD25, PD-1, Nrp1, ICOS, KLRG1, ST2, Ki67, Helios, T-bet, CTLA4, shown for blood and different brain regions. The adjusted P value reflects the cross-entropy difference between tSNE plots in brain region versus blood. (R) CD69 expression in Treg from the brain regions in pups and adult mice. (S) Heatmap showing expression of markers in brain region Treg. (T) Unaffected human brain tissues removed during brain surgery were compared to peripheral blood mononuclear cells by high-dimensional flow cytometry (n = 4). Representative histograms for CD4+Foxp3- T cells from human peripheral blood mononuclear cells, white matter, gray matter and meninges for CCR2, CXCR3, PD-1 and CD69. (U) Expression of ICOS, CD28, CD69, Ki-67, CD95, CD31, HLA-DR, CCR2, CXCR5, CD25, PD-1, CXCR3, RORγT, CCR4, CTLA-4, CCR7 and CD45RA on CD4+Foxp3- T cells. (V) tSNE of CD4+Foxp3+ T cells gated on CD4+Foxp3+CD3+CD8-CD14- cells and built on ICOS, CD28, CD69, Ki-67, CD95, CD31, HLA-DR, CCR2, CXCR5, CD25, PD-1, CXCR3, RORγT, CCR4, CTLA-4, CCR7 and CD45RA. Colors indicate FlowSOM clusters. Dendrogram showing the relationship across the brain regions based on cross-entropy differences in tSNE. (W) Representative histograms for CD4+Foxp3+ T cells from human peripheral blood mononuclear cells, white matter, gray matter and meninges for CCR2, CXCR3, PD-1 and CD69. (X) Expression of ICOS, CD28, CD69, Ki-67, CD95, CD31, HLA-DR, CCR2, CXCR5, CD25, PD-1, CXCR3, RORγT, CCR4, CTLA-4, CCR7 and CD45RA on CD4+Foxp3+ T cells. (Y) Single cell RNaseq data from sorted CD4+CD3+CD8- cells from the human brain and PBMCs. Quality control filtering and gating based on expression markers identified 86 CD4+ T cells from the brain and 567 CD4+ T cells from the blood. tSNE dimensionality reduction is used for cluster display, with lineage marker expression indicated by color for CD3D, CD4, IL7R, IL2RA, FOXP3, CD44, SELL, AREG, CD69, KLRG1 and NR4A1. (Z) Differentially expressed genes were assessed for pathway by GSEA.