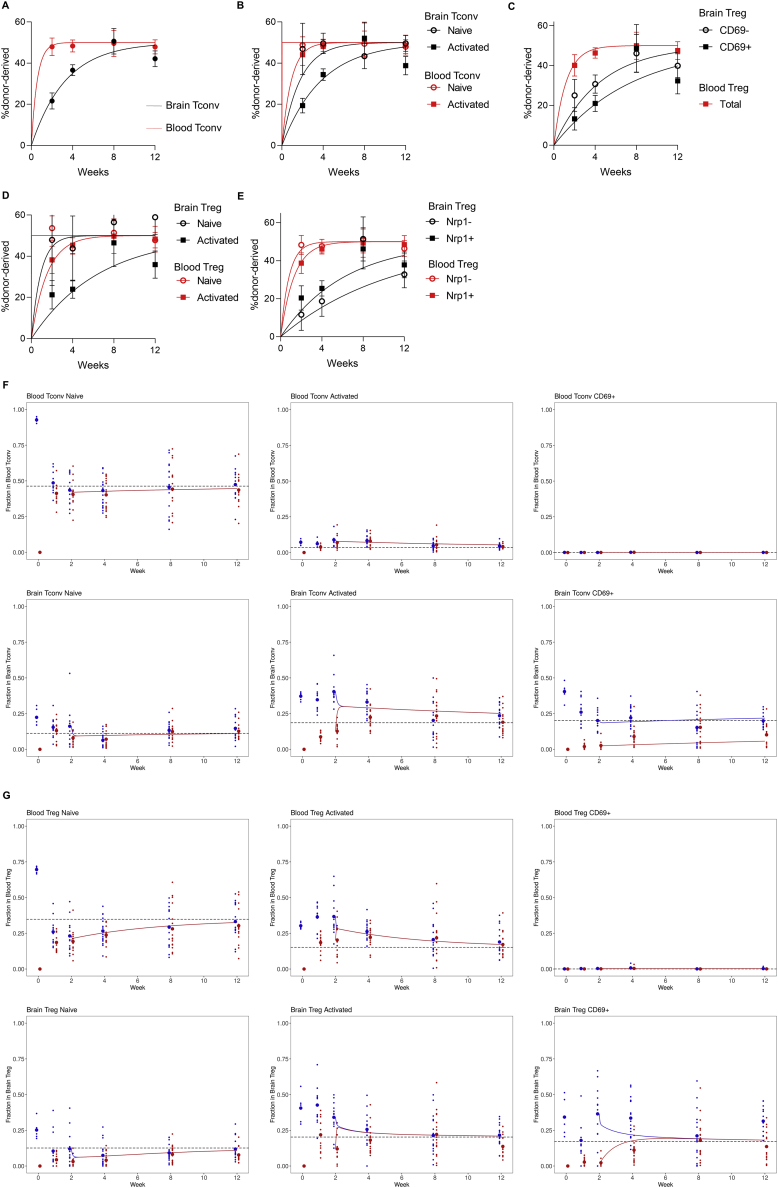
Figure S3.
Parabiotic Analysis of Brain T Cell Kinetics, Related to Figure 3
Parabiosis surgery was performed on Foxp3Thy1.1CD45.1 and Foxp3Thy1.1CD45.2 mice, with analysis of brain and blood CD4 T cells at 1, 2, 4, 8 and 12 weeks (n = 12, 12, 18, 16, 14). Curves of best fit as well as the mean ± SEM for each subset are displayed at each time-point. (A) Curve of best fit for the origin of CD4+Foxp3- conventional T cells in the blood and brain. (B) Curve of best fit for the origin of CD4+Foxp3- conventional T cells in the blood and brain, divided into naive (CD62LhiCD44low) and antigen-experienced (CD62LlowCD44hi) subsets. (C) Curve of best fit for the origin of CD4+Foxp3+ regulatory T cells in the blood and brain, divided into CD69- and CD69+ subsets in the brain. (D) Curve of best fit for the origin of CD4+Foxp3+ regulatory T cells in the blood and brain, divided into naive (CD62LhiCD44low) and antigen-experienced (CD62LlowCD44hi) subsets. (E) Curve of best fit for the origin of CD4+Foxp3+ regulatory T cells in the blood and brain, divided into thymic-derived (Nrp1+) and peripherally-derived (Nrp1-) subsets. (F) Time evolution of average populations as predicted by a continuous-time Markov chain model fitted on the data from weeks 2 to 12, with homeostatic state identified from week 0. Experimental data (points) and calculations (solid lines) are shown in blue for host cells and in red for donor cells. Big points identify experimental population averages, whereas homeostatic states are displayed with black dashed lines. Model for CD4+Foxp3- conventional T cells and (G) CD4+Foxp3+ regulatory T cells. Tissue and subpopulation for each graph are shown in the top-left graph label.