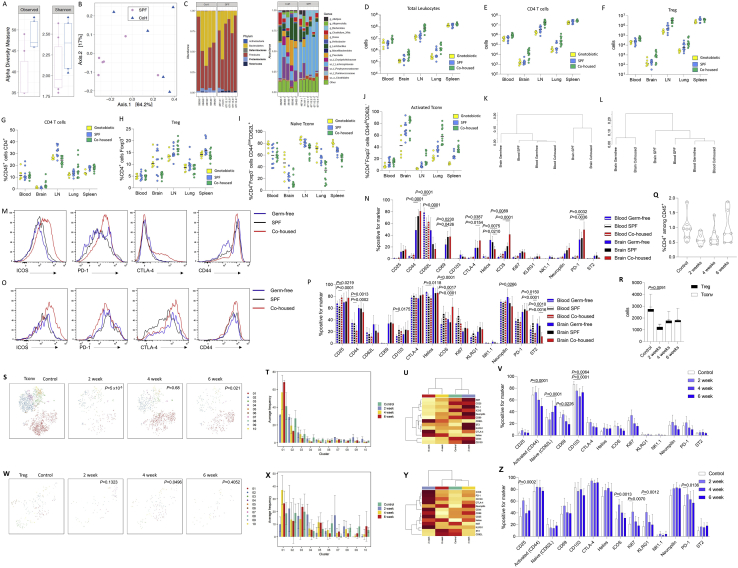
Figure S5.
Altered Activated Conventional Brain CD4 T Cells following Exposure to a Wild Microbiome or Broad-Spectrum Antibiotic Treatment, Related to Figure 5
(A) The microbial diversity of caecal contents in specific pathogen-free (SPF) and wild-cohoused (CoH) mice was compared through 16S rRNA analysis. When comparing the microbial communities from SPE and CoH mice groups, we detect differences in richness but not in the Shannon diversity index (p < 0.03 and p > 0.05, respectively, Wilcoxon test) in the CoH compared to SPF mice. 32% of the variation on the microbial composition can be explained by this grouping (adonis test, R2 = 0.31681, p = 0.022). (B) Principal coordinate analysis (PCoA) of the mice microbiota community variation based on Bacteria and Archaea genus-level Bray-Curtis dissimilarity distances revealed two different clusters. (C) Phylum and genus distribution barplots. Only the top 14 genera are displayed. The body of the boxplot represents the first and third quartiles of the distribution and the median line. The whiskers extend from the quartiles to the last data point within 1.5 × IQR, with outliers beyond. ‘uc_f’: unclassified genus of the family and ‘uc_o’: unclassified genus of the order. Differential abundance analysis at the phylum level identified Deferribacteres and Proteobacteria enriched and Tenericutes depleted in CoH mice (q-values < 0.1, Wilcoxon test). (D) Lymph nodes, spleen, blood, perfused brain and perfused lung were dissected from gnotobiotic, SPF and dirty cohoused mice (n = 6, 10, 12) and assessed by flow cytometry. Total leukocyte counts, and (E) absolute numbers of CD4 T cells or (F) Tregs. (G) Proportion of CD4 T cells within the CD45+ compartment. (H) Proportion of Tregs within the CD4+ compartment. (I) Proportion of naive or (J) activated cells within the conventional T cell compartment. (K) Dendrogram showing the relationship of conventional T cells and (L) Tregs across the samples, based on cross-entropy differences in tSNE for expression of CD62L, CD44, CD103, CD69, CD25, PD-1, Nrp1, ICOS, KLRG1, ST2, Ki67, Helios, T-bet, CTLA4. (M) Representative histograms shown for ICOS, PD-1, CTLA-4 and CD44 on conventional CD4 T cells. (N) Expression of assessed markers on conventional CD4 T cells. (O) Representative histograms shown for ICOS, PD-1, CTLA-4 and CD44 on Tregs. (P) Expression of assessed markers on Tregs. (Q) Wild-type mice were placed on broad-spectrum antibiotics for 2, 4 or 6 weeks (n = 5, 8, 6) and were compared to untreated control mice (n = 8) by high parameter flow cytometry of the perfused brain. CD4 T cells in perfused brain, as a percentage of CD45+ cells, and (R) absolute numbers of conventional and regulatory CD4 T cells in the brain. P value refers to comparison of conventional T cells. (S) tSNE of CD4+Foxp3- T cells gated on CD4+Foxp3-CD3+CD8-CD45+ cells and built on CD62L, CD44, CD103, CD69, CD25, PD-1, Nrp1, ICOS, KLRG1, ST2, Ki67, Helios, T-bet, CTLA4. P values represent cross-entropy comparison to control mice. FlowSOM clusters are illustrated in color and (T) quantified. (U) Expression heatmap and (V) marker expression rate for CD62L, CD44, CD103, CD69, CD25, PD-1, Nrp1, ICOS, KLRG1, ST2, Ki67, Helios, T-bet and CTLA4 levels in CD4+Foxp3- T cells. (W) tSNE of CD4+Foxp3+ T cells gated on CD4+Foxp3+CD3+CD8-CD45+ cells and built on CD62L, CD44, CD103, CD69, CD25, PD-1, Nrp1, ICOS, KLRG1, ST2, Ki67, Helios, T-bet, CTLA4. P values represent cross-entropy comparison to control mice. FlowSOM clusters are illustrated in color and (X) quantified. (Y) Expression heatmap and (Z) marker expression rate for CD62L, CD44, CD103, CD69, CD25, PD-1, Nrp1, ICOS, KLRG1, ST2, Ki67, Helios, T-bet and CTLA4 levels in CD4+Foxp3+ T cells.