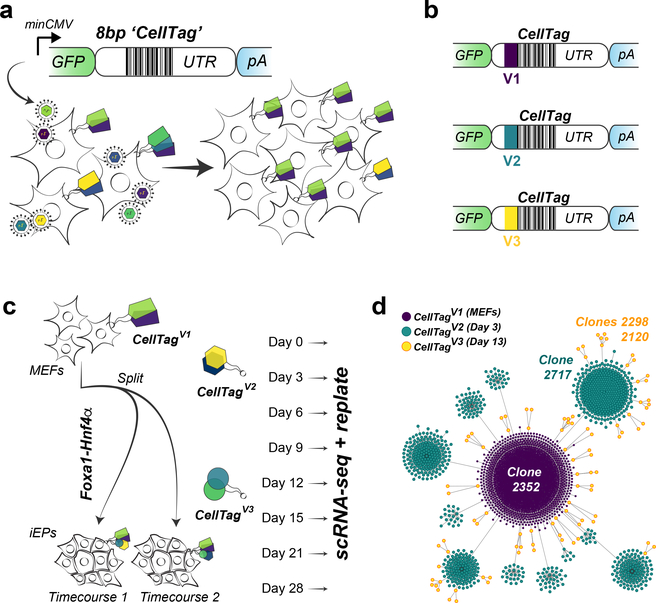
Fig. 1 |. The CellTag workflow for parallel capture of lineage and identity.
a, Schematic of the CellTagging workflow. A lentiviral construct is engineered with an 8-bp random ‘CellTag’ barcode in the 3′ UTR of GFP, followed by an SV40 polyadenylation signal. Cells are transduced with this lentiviral library (produced via transfection of HEK293T cells with the complex plasmid library) so that each cell expresses ~3–4 CellTags, resulting in a unique, heritable signature, enabling clonally related cells to be tracked over the course of an experiment. b, Design of CellTag constructs for multiplexing: a short, 6-bp index sequence is inserted in front of the variable CellTag region, allowing different rounds of CellTagging to be demultiplexed and lineage trees to be subsequently constructed. c, Schematic of experimental approach: reprogramming of MEFs to iEPs via retroviral delivery of Foxa1 and Hnf4α. Cells were transduced as fibroblasts with CellTagV1, and again at 3 d (with CellTagV2) and 13 d (with CellTagV3) after initiation of reprogramming. A fraction of cells were recovered for scRNA-seq every 3–7 d, and the remainder were replated. d, Reconstruction and visualization of lineages via force-directed graphing. Each node represents an individual cell, and edges represent clonal relationships between cells: purple, CellTagV1 clones; blue, CellTagV2 clones; yellow, CellTagV3 clones. n = 2,199 cells.