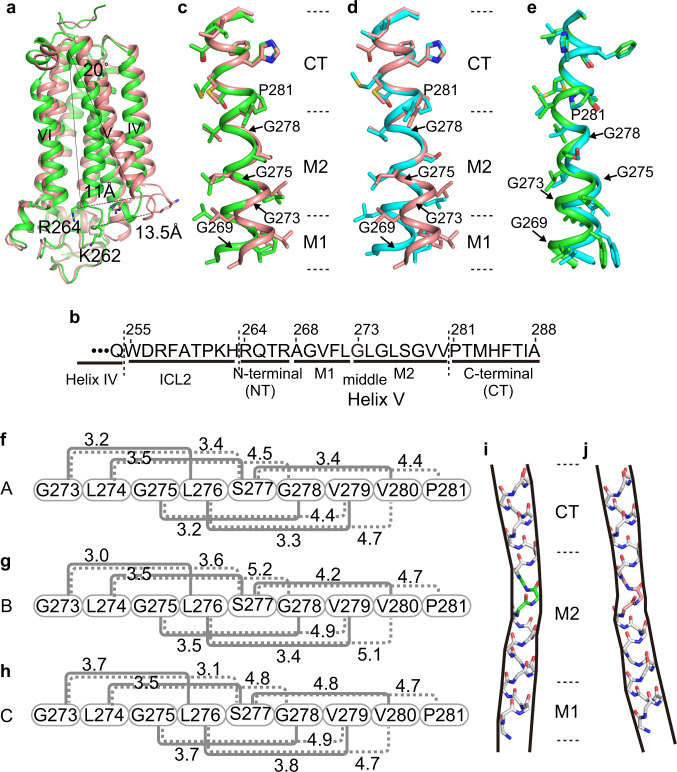
Fig. 2. Structures of helix V in the closed and open forms of AdipoR1(A208).
a Superimposition of the closed-form (molecule A, green) and open-form (molecule C, salmon) structures. The cytoplasmic ends of helices V are shifted by 11 Å between the two structures. b The amino acid sequences of ICL2 (Trp255–His263) and helix V (Arg264–Ala288), further divided into the N-terminal (NT) region (Arg264–Arg276), the middle (M1 and M2) regions (Ala268–Leu272 and G273–Val280, respectively), and the C-terminal (CT) region (Pro281–Ala288), of AdipoR1. c–e Structure comparison along the M1, M2, and CT regions of helix V among molecules A (green), B (cyan), and C (salmon), by superimposition of the CT region. f–h The distances (Å) of the main-chain amide carbonyl oxygen atom (CO) of residue i with the main-chain amide nitrogen atom (HN) of residues i + 3 (solid gray line) and i + 4 (dashed gray line) in the M2 region (i = 273–277) of helix V of molecules A (f), B (g), and C (h). According to these CO–HN distances and the bond angles in the M2 region (Supplementary Table 1), a 310-helical conformation, rather than an α-helix, is assumed with four weak hydrogen bonds (3.2–3.5 Å) between the COs of Leu274–Ser277 (residues i) and the HNs of Ser277–Val280 (residues i + 3) in molecule A (f), whereas such 310-helical hydrogen bonding is weaker in molecule B (g) and almost negligible in molecule C (h). The CO of Gly273 does not form a hydrogen bond in molecules A–C (Supplementary Table 1). i, j Schematic representations of the main chain of helix V, which is bent inward in molecule A (i) and rather straight in molecule C (j). The 310-helical conformations of Leu276–Ser277–Gly278, colored green in molecule A (i) and salmon in molecule C (j), in the center of the M2 region are thinner than the α-helical conformations in the M1 and CT regions.