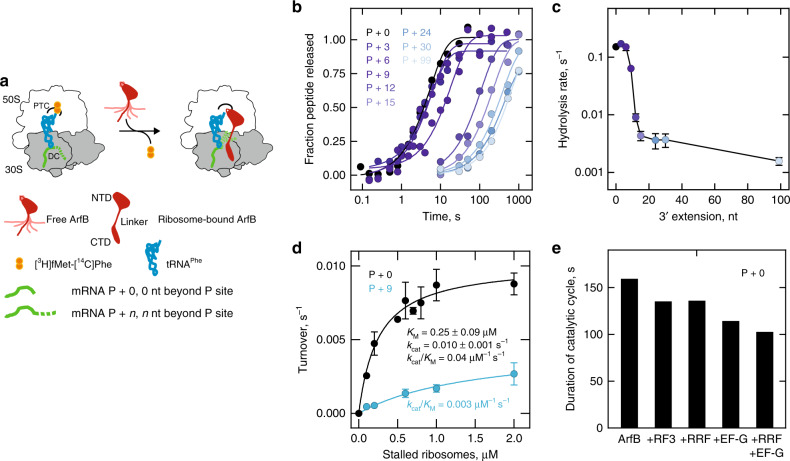
Fig. 1. ArfB preferentially rescues ribosomes stalled on short mRNA.
a Experimental assay for ArfB-mediated ribosome rescue. Ribosome complexes with mRNAs of different length are rapidly mixed with ArfB and the fraction of peptides released from tRNA by ArfB-dependent hydrolysis is quantified. PTC peptidyl transferase center; DC decoding center; NTD N-terminal domain; CTD C-terminal domain. Free ArfB is shown as an ensemble of dynamic molecules10, the ribosome-bound ArfB is shown as in the X-ray structure15. b Time courses of single-round peptidyl-tRNA hydrolysis at excess ArfB (1 µM) over ribosome complexes (0.15 µM). The mRNA length is indicated by number of nucleotides (nt) extending beyond the P site, from none (P + 0) to 99 nt (P + 99). Data represented as mean values of two biological replicates. Solid lines are exponential fits. c Rate of hydrolysis at increasing mRNA length. Error bars indicate the SEM of the exponential fits (b). d Peptidyl-tRNA hydrolysis on P + 0 and P + 9 complexes at limiting ArfB concentrations. Initial velocity of the hydrolysis reaction is measured after mixing ArfB (0.02 µM) with increasing concentrations of P + 0 or P + 9 complexes. Solid lines are results of hyperbolic fitting. Error bars represent the SEM of three biological replicates. e Effect of ribosome recycling factors on the duration of an ArfB catalytic cycle on P + 0 complexes (0.2 µM) mixed with catalytic amounts of ArfB (0.02 µM) and excess of RF3 (0.5 µM), RRF (0.5 µM) and EF-G (0.5 µM). All experiments were carried out at 37 °C.