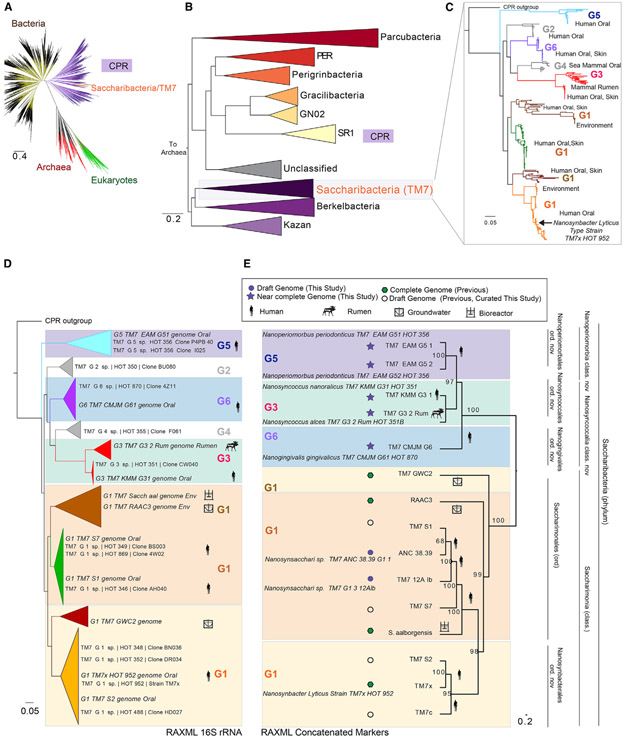
Figure 1. Expanded Coverage of the Saccharibacteria Phylum.
(A) Current view of the tree of life highlighting the Saccharibacteria and candidate phyla radiation (tree inferred from concatenated ribosomal gene dataset provided by Hug et al., 2016).
(B) Phylum-level maximum likelihood concatenated ribosomal gene tree of the CPR indicating that Saccharibacteria share a common ancestor with “Ca. Berkelbacteria” and “Ca .Kazan” groups.
(C) Fast tree 16S rRNA gene phylogeny of the major Saccharibacteria groups derived from public ribosomal databases and available genomes. The complete tree is available in a circular format with full bootstrap values in Figure S1 and in Newick format in Data S1.
(D and E) Phylogenetic relationships within the Saccharibacteria phylum using maximum likelihood 16S rRNA and concatenated ribosomal marker gene inference. (D) Maximum likelihood 16S rRNA gene phylogeny expanded tree, sequences from the Human Oral Microbiome Database, and those extracted from genomic assemblies are shown. (E) Concatenated ribosomal RAxML gene tree for new and previously published complete and draft genomes including first representative sequences from the G5 (Ca. Nanoperiomorbus periodonticus), G6 (Ca. Nanogingivalis gingivalicus), and G3 (Ca. Nanosyncoccus nanoralicus, Ca. Nanosyncoccus alces) lineages. Purple filled star and purple filled circle are assemblies from this study. Green filled hexagon and empty circle are assemblies from previous studies. Genomic data source material are indicated with icons. Genome properties and names for the provisionally proposed novel classes and orders are explained in Tables S1 and S2.