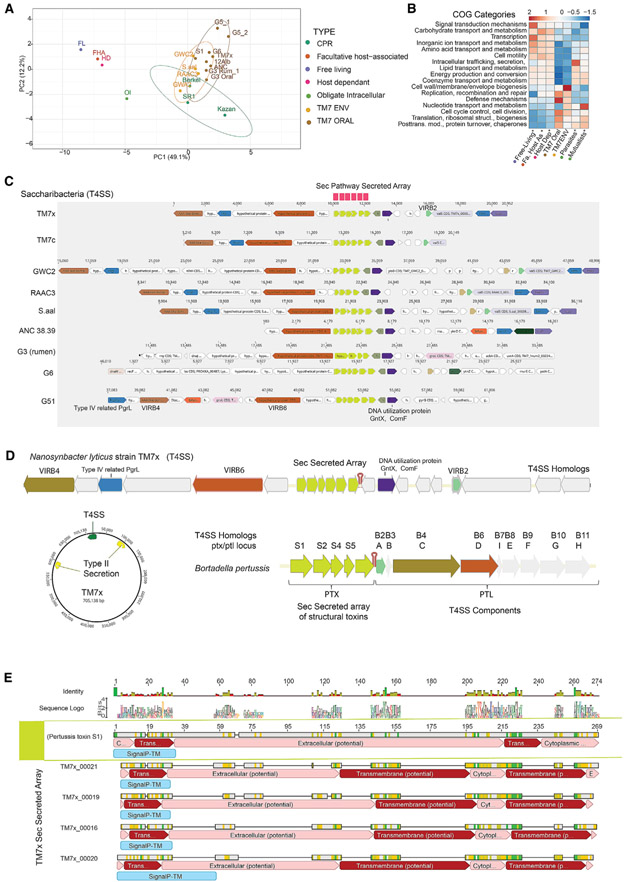
Figure 5. Comparative Analyses of Predicted Gene Functions and Conservation of T4SS with Novel Secreted Protein Array.
(A) Principal-component analysis (PCA) clustering of COGs distribution percentages averaged across oral and environmental Saccharibacteria groups for functionally annotated genes within free-living, facultative host-associated, and host-dependent as well as obligate intracellular (mutualistic and parasitic) bacteria.
(B) COGs distribution percentage across genomes for functionally annotated genes within these different lifestyle groups compared with environmental and oral Saccharibacteria groups (additional analyses in Table S8 and Figure S5). Rows are centered and unit variance scaled.
(C) Both environmental and mammalian-associated Saccharibacteria genomes maintained a region containing homologs of the type IV secretion system (VirB4, B6, and B2) and a novel array of small hypothetical proteins (4–6) containing N-terminal signal peptides targeting them for secretion through the sec pathway. This unique array of proteins was found across other reduced bacterial genomes and CPR (Figure S3).
(D) The arrangement of the six small secreted proteins in Nanosynbacter lyticus strain TM7x and T4SS homologs compared with known T4SS systems revealed similarities to the ptx/ptl locus of Bordetella pertussis. The effector proteins secreted by the Ptl machinery in B. pertussis form a large complex called the pertussis toxin (Ptx).
(E) Gapped alignment of pertussis toxin S1 protein with four of the six proteins in the TM7x array. The Ptx subunit, similar to the four TM7x proteins, has an N-terminal signal sequence and is translocated across the inner membrane by the general SecYEG secretory pathway.