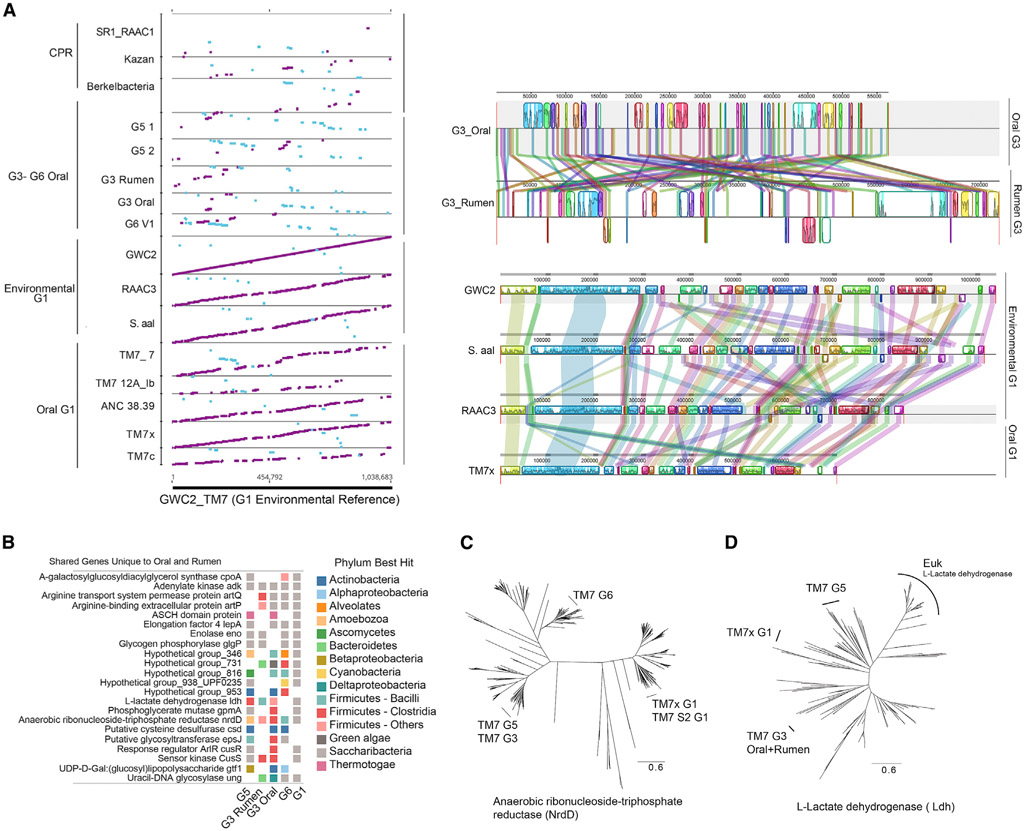
Figure 6. Mammalian Host and Environmental G1 Are Highly Conserved, and Shared Functions Are Independently Acquired in Mammalian Host-Associated Groups.
(A) All assembled contigs aligned against reference groundwater Saccharibacteria genome GWC2 reveals the synteny maintained across the environmental and human oral G1 group members. Large syntenic blocks are present and maintained in order when comparing full genomes of G1. In contrast, the G3 group containing human oral and rumen members has maintained smaller syntenic blocks with multiple re-arrangements.
(B) A total of 21 unique genes are shared with four or more of the oral genomes and are not found in environmental genomes, which indicates they may have been acquired during mammalian host adaptation. Colored squares indicate presence. Color indicates the phylum with the closest homolog. Several of the unique genes that were potentially acquired show mixed taxonomy across groups, indicating convergent evolution by horizontal gene transfer.
(C) Maximum likelihood phylogenetic protein tree of anaerobic ribonucleoside-triphosphate reductase (NrdD) displaying divergent phylogenetic relatedness among Saccharibacteria groups (full trees in Figure S6).
(D) The L-lactate dehydrogenase gene was identified as being a unique gene among the mammalian associated groups that was not present in the environmental genomes. The taxonomic best hits in (B) and the maximum likelihood protein tree shown here indicate divergent phylogenetic relatedness between the acquired genes, supporting independent acquisition from different bacteria.