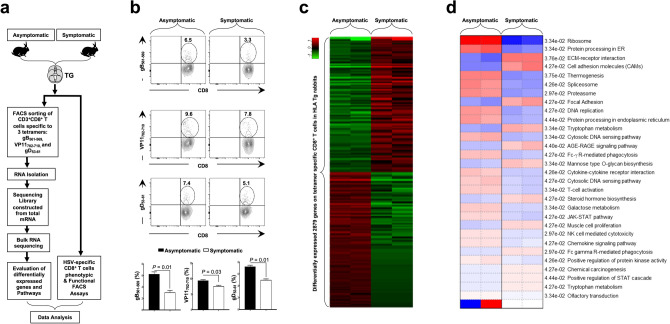
Figure 4.
Differential gene expression in HSV-specific CD8+ T cells from trigeminal ganglia of HSV-1 infected symptomatic vs. asymptomatic HLA Tg rabbits. (a) Experimental design and validation of differentially expressed genes in CD8+ T cells sharing the same HSV-1 epitope-specificities, from SYMP and ASYMP HLA Tg rabbits. CD8+ T cells specific to HLA-A*0201-restricted HSV-1 gB561–567, VP11/12702–710, and gD53–61 epitopes were sorted from TG of HLA-A*0201-positive SYMP and ASYMP HLA Tg rabbits, using specific tetramers. Total RNA was extracted from each clone of epitope-specific CD8+ T cells, and whole transcriptome analysis was performed using bulk RNA sequencing to determine the levels of expression of 23,669 rabbit genes (OryCun2.0 (GCA_000003625.1). (b) Frequencies of CD8+ T cells specific to HLA-A*0201-restricted HSV-1 gB561–567, VP11/12702–710, and gD53–61 epitopes detected by FACS in TG of HLA-Tg rabbits. (c) The heatmap is showing the most significant 2,879 differentially expressed genes among SYMP and ASYMP HLA Tg rabbits. Genes with minimum count per million (CPM) ≥ 0.5 were used for obtaining the transformed counts data for clustering using regularized log (rlog). (d) Bulk RNA heatmap shows the pathways that are different among ASYMP and SYMP HLA Tg rabbits. Genes differentially expressed in both single-cell RNA sequencing and bulk RNA sequencing were considered for pathway analyses. Parametric gene set enrichment analysis (PSGEA) method based on data curated in Gene Ontology and KEGG was applied. Pathway significance cut-off with a false discovery date (FDR) ≥ 0.2 was applied.