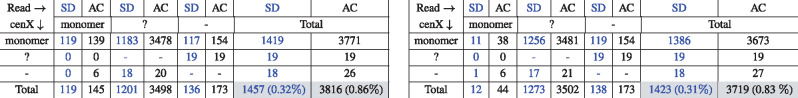
Table 1.
Summary of errors in the monoread-to-monocentromere alignments computed by the AC (black) and SD (blue) tools for 12 monomers (Left) and for 13 monomers that include 12 known cenX monomers and a novel (K + F) monomer (Right)
|
Notes: Symbol ≪monomer≫ corresponds to 1 of the 12 cenX monomers or (K + F) monomer, ≪?≫ corresponds to a gap symbol, ≪-≫ corresponds to a space symbol representing an indel in alignment of mono(Read) against . A cell (i, j) represents the number of times when a symbol of type i in was aligned to a symbol of type j in mono(Read). For 12 (13) monomers decomposition, the number of matches is 445 169 (445 206) for SD and 442 783 (442 887) for AC.
