Fig. 5. 5′ and 3′ 12-RSS utilization in VHQ52.2.4 or VH7183.4.6-DH recombination.
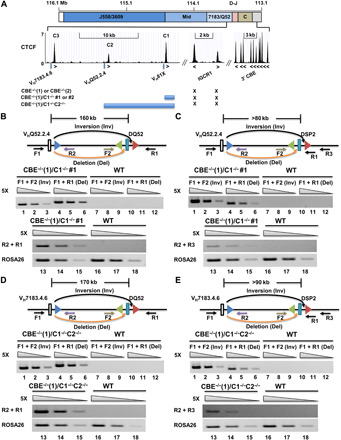
(A) Schematic of the 3′ IgH locus CTCF-binding sites (24) and mutations produced by CRISPR-Cas9 in the context of the IGCR1 mutated cell line CBE−/−(1). (B and D) Rearrangements assays of VHQ52.2.4 (B) or VH7183.4.6 (D) to 5′- or 3′-RSS of DQ52 by deletion (orange arrows) or inversion (black arrows), respectively. Locations and orientation of primers used to assay recombination are indicated, together with the 23-RSS of VHQ52.2.4 (B) or VH7183.4.6 (D) (blue triangles) and 12-RSSs flanking DQ52 gene segments (green and red triangles). Each set of three lanes contains fivefold increasing amounts of genomic DNA starting at 8 ng (lanes 3, 6, 9, 12, 15, and 18). ROSA26 was used as the loading control. Data shown are representative of two biological replicate experiments. (C and E) Rearrangements assays of VHQ52.2.4 (B) or VH7183.4.6 (D) to 5′- or 3′-RSS of DSP2 by deletion or inversion, respectively. ROSA26 was used as the loading control. Data shown are representative of two biological replicate experiments.
