Figure 3:
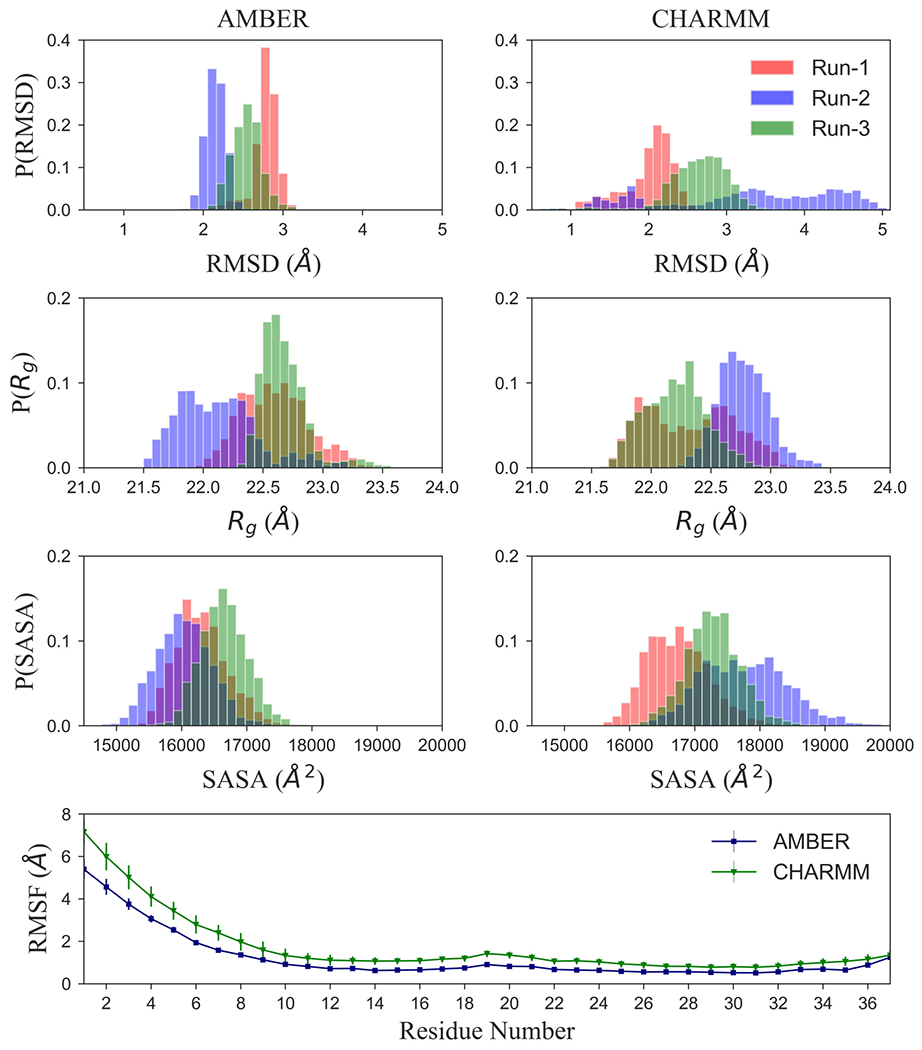
Comparison of molecular dynamics simulations of L-amylin fibril fragment using either AMBER ff99SB or CHARMM36. For this purpose, we show in (a) the normalized distribution of the root-mean-square deviation (RMSD), (b) the radius of Gyration (Rg), and (c) of the solvent-accessible-surface-area (SASA). In (d), we show the root-mean-square-fluctuation (RMSF) of the Cα atoms. For RMSF, values are averaged over all chains in the fibril fragment and all three trajectories.
