FIGURE 1.
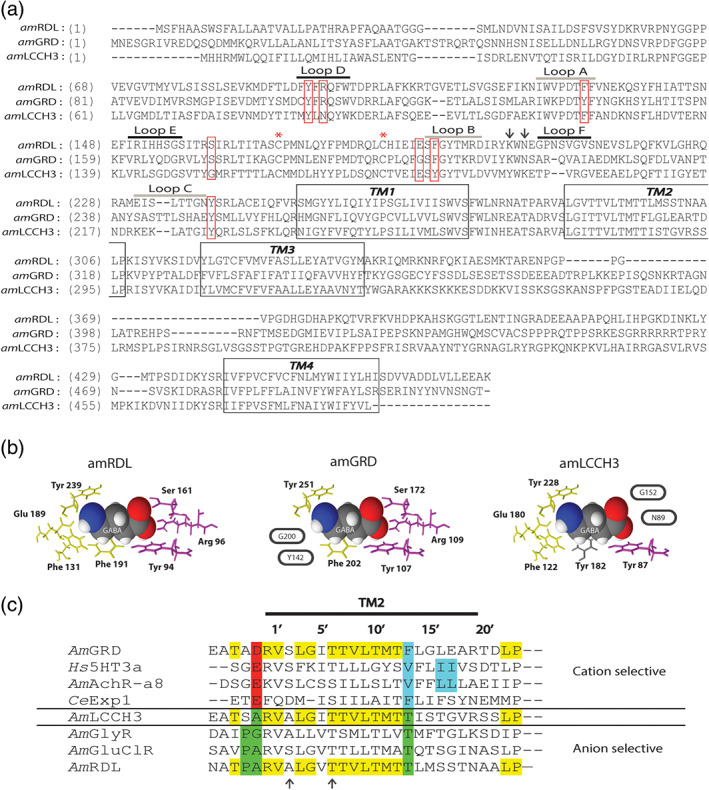
AmGABA receptor sequence analysis. (a) Alignment of honeybee RDL, GRD, and LCCH3 subunit polypeptidic sequences. The four transmembrane segments are marked by black boxes. The two asterisks correspond to the two cysteines involved in the stereotypical motif “Cys‐loop.” The pentameric structure and organization of insect GABA receptors are assumed from mammalian GABA receptors. On this basis, the agonist pocket would be located at the interface between two successive subunits, the principal and the complementary subunits. The six extracellular loops structuring the agonist pocket are underlined above the sequence. Loops A to C (grey line) are on the principal subunit, while loops D to F (black line) are on the complementary subunit. The seven amino acids assumed to directly interact with the GABA molecule are indicated by red boxes. The two arrows locate the charged lysine residues implicated in first events of GABA binding. (b) Compacted representation of a GABA molecule surrounded by its seven interacting residues in the GABA binding pocket of each honeybee GABA receptor subunit (inspired from Comitani, Limongelli, & Molteni, 2016). The GABA binding site is located at the interface of two GABA receptor subunits that both participate in shaping a GABA binding site. The first part of this binding site is made of four amino acids of the principal subunit (in yellow), and the second part is made of three amino acids of the complementary subunit (in purple). The GABA binding site is fully conserved in AmRDL and allows homomeric RDL assembly to exhibit functional GABA binding sites. Two amino acids of AmGRD as principal subunit and two amino acids of AmLCCH3 as complementary subunit are non‐conserved (circled), which probably alters the functionality of these half binding sites and produce homomeric GRD or LCCH3 receptors unable to make functional GABA binding sites. The phenylalanine of loop B in the typical GABA binding site is also not conserved in AmLCCH3, but the functionality of this phenylalanine could be preserved by the presence of a tyrosine residue (Y182) at the same position (coloured in grey). (c) Alignment of the pore forming TM2 segment of Apis mellifera RDL, LCCH3, GRD, glycine receptor (GlyR), nicotinic receptor (AchRα8), glutamate‐gated chloride channel (GluClR), Homo sapiens 5‐HT receptor (5HT3α) and Caenorhabditis elegans Exp‐1 receptor (from Gisselmann et al., 2004). The conserved amino acids in the three sequences AmRDL, AmGRD, and AmLCCH3 are in yellow. AmRDL possesses the key elements of anionic conductance coloured in green, with proline in position −2′ and threonine in position 13′. AmGRD possesses a negatively charged amino acid in position −1′, an aspartic acid, which is a hallmark of cationic conductance (in red). Amino acids crucial for the calcium permeability of cationic receptor are in blue. A mutation of one of the two successive leucine residues in human AchRα7 abolishes calcium permeability. Arrows highlight amino acids important for dieldrin sensitivity. The threonine in position 6′ is conserved in all AmGABA receptor subunits, but the alanine in position 2′ is absent in AmGRD
