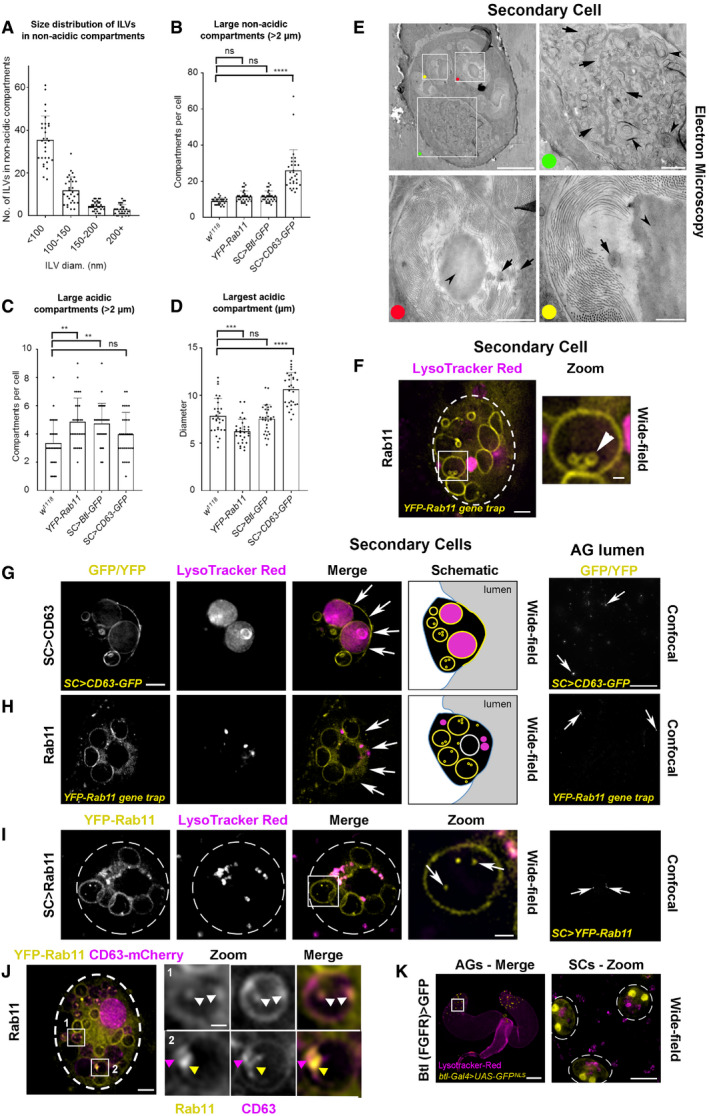
Wide‐field fluorescence images of living fly secondary cells (SCs) and paired accessory glands (AGs; K), and confocal images of fixed AG lumens (G–I). Focal planes indicated in Fig
1A schematics. Acidic compartments are marked by the vital dye LysoTracker
® Red (magenta); GFP‐ and YFP‐tagged constructs are shown in yellow. (F) Basal view of an SC expressing a
YFP‐Rab11 gene trap, with cell outline approximated by dashed white circle. Rab11 compartment highlighted by box contains three large ILVs (arrowhead) with Rab11 marking their outlines. (G) Transverse view of an SC from a fly in which CD63‐GFP is expressed specifically in SCs under GAL4/UAS control. CD63‐GFP is present on both the limiting and ILV membranes of acidic and non‐acidic compartments, in addition to the apical plasma membrane (arrows in merge). Image is also shown schematically. CD63‐GFP puncta (arrows) are present in the AG lumen of these flies. (H) Transverse view of an SC expressing a
YFP‐Rab11 gene trap, which marks non‐acidic compartments, and also the cytosol, but is not trafficked to the apical plasma membrane (arrows in merge). Image is also shown schematically. Absence of YFP‐Rab11 throughout the apical plasma membrane is shown in the Z‐stack in
Movie EV2. YFP‐Rab11 puncta (arrows) are present at low levels in the AG lumen. (I) Basal view of an SC from a fly expressing SC‐specific YFP‐Rab11 (yellow) under GAL4/UAS control, with cell outline approximated by dashed white circle. Boxed non‐acidic compartment is magnified in Zoom; arrows highlight YFP‐Rab11‐positive ILVs (Merge) and puncta present at low levels (AG lumen). (J) Basal view of an SC from a fly expressing the
YFP‐Rab11 gene trap (yellow) and SC‐specific CD63‐mCherry (magenta) under GAL4/UAS control, with cell outline approximated by dashed white circle. Large non‐acidic compartments with CD63‐mCherry at their limiting membrane appear to exclude YFP‐Rab11 from their surface (boxes 1 and 2). In the compartment lumen, co‐localisation of the markers is observed (white arrowheads; box 1), but also non‐overlapping fluorescence (coloured arrowheads; box 2), consistent with ILV heterogeneity in a single compartment. (K) AGs from a fly expressing nuclear GFP under the control of a well‐characterised
btl‐GAL4 enhancer trap (Hayashi
et al,
2002). Boxed region of AG epithelium containing SCs is enlarged in Zoom, with SC outlines approximated by dashed white circles. GFP localisation in binucleate SCs suggests that
btl is normally expressed in SCs.