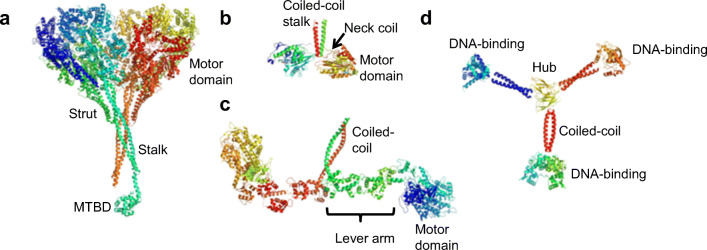
Fig. 1.
The structures of linear protein motors drawn to scale and oriented so that the track would be at the bottom of the figure. a Crystal structure of Dictyostelium discoideum dynein dimer determined at a 3.8-Å resolution (PDB 3VKH) with the motor domain (top) distal from the microtubule-binding domain (MTBD). Only one of the two MTBDs was resolved in the crystal structure (Kon et al. 2012). b The crystal structure of the kinesin dimer (PDB 3KIN) from rat brain determined at a 3.1-Å resolution (Kozielski et al. 1997). The arrow shows the neck coil linker (red line) leading from the right-hand motor domain to the C-terminal coiled-coil helix (red spiral). The green line near the arrowhead is the N-terminus of the right-hand motor domain. The motor domain binds directly to the microtubule track. c The structure of chicken smooth muscle heavy meromyosin dimer (PDB 3J04) determined by electron crystallography at a 20-Å resolution (Baumann et al. 2012). The two myosin heads splay out from the C-terminal coiled-coil. One lever arm and motor domain are labelled. The motor domains bind directly to the actin track. d A model for the tumbleweed artificial clocked walker protein (Bromley et al. 2009). The tumbleweed has three ligand-gated DNA-binding domains that are joined to a central hub via coiled-coil arms