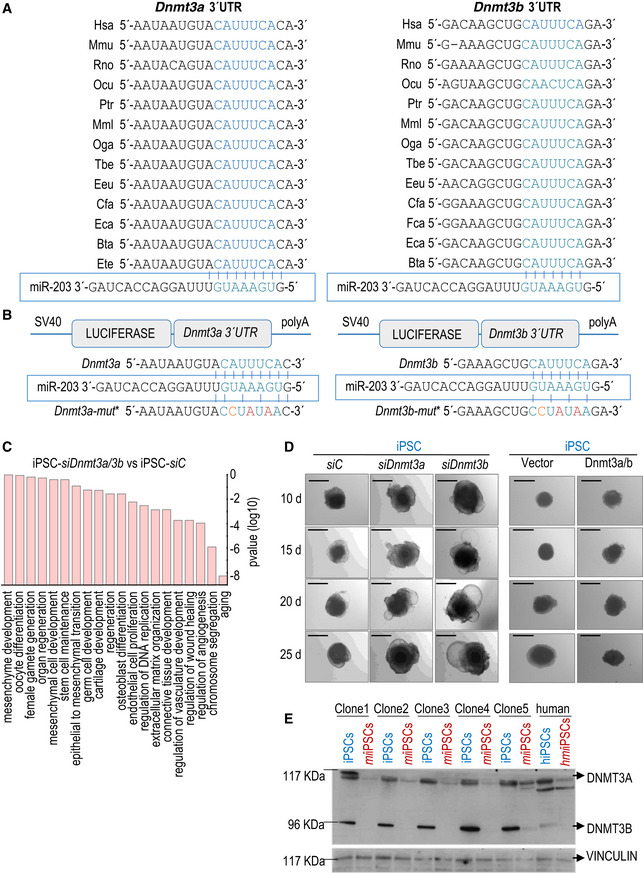
Dnmt3a and Dnmt3b 3′‐UTR alignment in several representative species. The seed region of the miR‐203 target site contained in these 3′‐UTRs is highlighted in blue and aligned with the corresponding miR‐203 seed sequence.
Schematic representation of the luciferase reporter, carrying the wild‐type Dnmt3a (left) or Dnmt3b (right) complete 3′‐UTRs or the corresponding mutated versions, downstream of the luciferase gene. The seed region of the miR‐203 target site contained in these 3′‐UTRs is written in blue and aligned with the corresponding miR‐203 seed sequence. The mutated residues are indicated in red.
Major pathways from the Gene Ontology database upregulated in wild‐type iPSCs treated with specific siRNAs against Dnmt3a and Dnmt3b versus scrambled sequences (siC). P‐values were calculated by Fisher's exact test.
Representative images of embryoid bodies (EBs) generated from wild‐type iPSCs treated with specific siRNAs either against Dnmt3a or Dnmt3b (left panel) or transfected with a combination of miR‐203‐resistant Dnmt3a/b cDNAs (right panel) at the indicated time points during the differentiation process. Scale bars, 500 μm.
Detection of Dnmt3a (117 KDa) and Dnmt3b (96 KDa) protein levels by Western blot, in 5 independent clones of mouse un‐induced iPSCs or miiPSCs and one human cell line, transfected with mimics control (hiPSCs) or miR‐203 mimics (hmiiPSCs). Protein extracts were evaluated immediately after miR‐203 exposure, to detect the modulation of Dnmt3a/b protein levels. Vinculin levels are included as loading control.