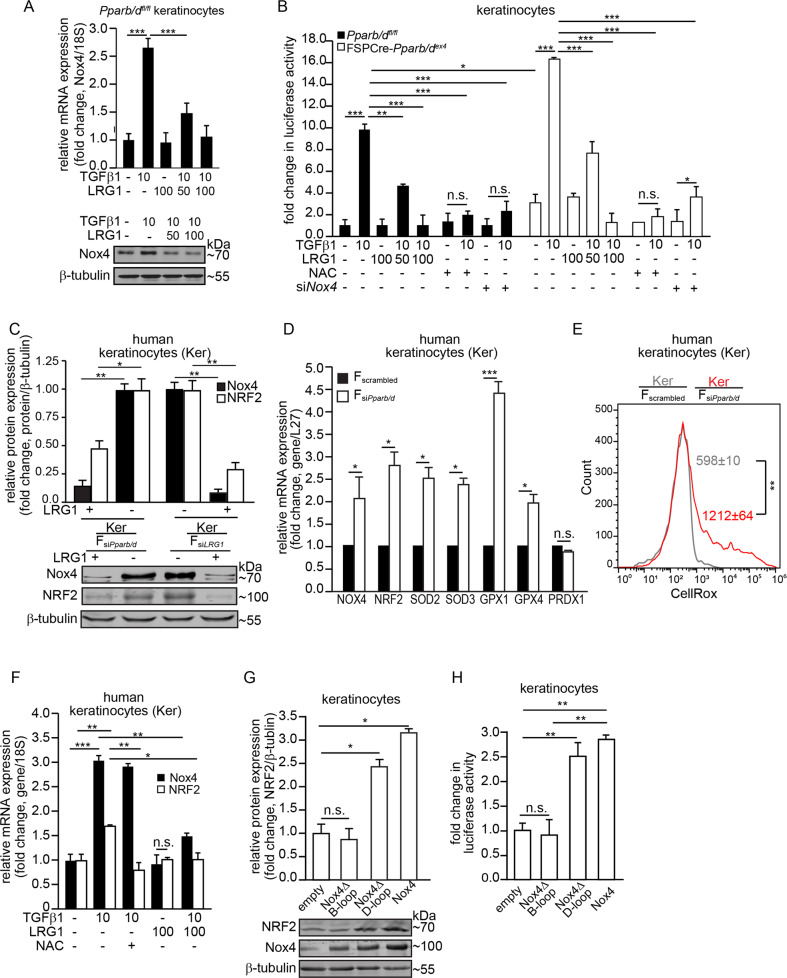
Fig. 4. Fibroblast PPARβ/δ modulates epidermal Nox4 via LRG1-TGFβ1 signaling.
a Relative mRNA (top panel) and protein (bottom panel) levels of Nox4 in Pparb/dfl/fl keratinocytes treated with either TGFβ1 or LRG1 proteins. Representative immunoblot for Nox4 is shown. β-tubulin served as a housekeeping protein from the same samples. A densitometric quantification plot is shown. b Fold change in ARE-dependent luciferase activities in FSPCre-Pparb/dex4 and Pparb/dfl/fl keratinocytes treated with either TGFβ1 (10 ng/mL) or LRG1 (50–100 ng/mL). Keratinocytes whose endogenous Nox4 expression was suppressed by siRNA were denoted by siNox4. Antioxidant NAC was used at 500 µM. For the reporter assay, firefly luciferase activity was normalized to Renilla luciferase activity. c Representative immunoblots of Nox4 and NRF2 in human keratinocytes (Kers) cocultured with either dermal human fibroblasts knockdown of Pparb/d (FsiPparb/d) or LRG1 (FsiLRG1) (bottom panel). β-tubulin served as housekeeping protein and was from the same samples. A densitometric quantification plot is shown (top panel). d Relative mRNA levels of the indicated antioxidant genes in Kers cocultured with FsiPparb/d or Fscrambled. Ribosomal protein L27 was used as a housekeeping gene. e Mean intracellular ROS level in Kers cocultured with either FsiPparb/d (red) or Fscrambled (gray). The histogram shows mean CellRox fluorescence readings in keratinocytes. f Relative mRNA levels of Nox4 and NRF2 in Kers treated with either TGFβ1, LRG1, or NAC. Ribosomal 18S was used as a housekeeping gene. g Representative immunoblots of Nox4 and NRF2 in primary wild-type mouse keratinocytes transfected with various Nox4 mutants. Densitometric quantification plots are shown (right panel). β-tubulin served as a housekeeping protein from the same samples. h Fold change in ARE-dependent luciferase activities in mouse keratinocytes overexpressing wild-type, Nox4ΔD-loop, and Nox4ΔB-loop Nox4 mutants. Empty vector served as the control. For the reporter assay, firefly luciferase activity was normalized to Renilla luciferase activity. All data are represented as the mean ± s.d. For a and e, n = 5 independent experiments. For b–d and f–h, n = 3 independent experiments. **p < 0.01.