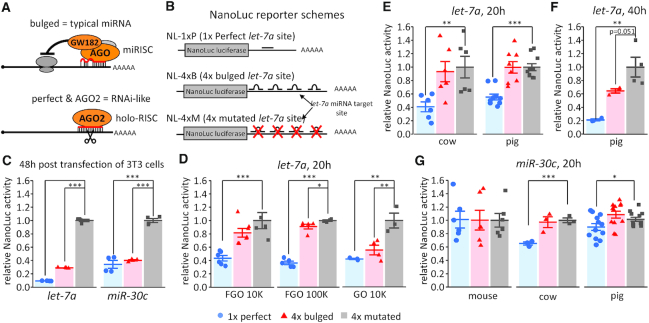
Figure 1.
Analysis of miRNA activity in mammalian oocytes with NanoLuc reporters. (A) Schematic difference between ‘bulged’ and ‘perfect’ miRNA binding sites. Bulged miRNA binding sites, which are typical of animal miRNAs, have imperfect complementarity and lead to translational repression followed by deadenylation and mRNA degradation (1). Perfect complementarity of miRNAs loaded on AGO2 results in RNAi-like endonucleolytic cleavage. (B) Schematic depiction of let-7a nanoluciferase reporter constructs used in the study. The miRNA sites were cloned in the 3′UTR either as a 1×-perfectly complementary site, a 4× site producing a bulged conformation or a 4× mutated site. An analogous set of reporters was constructed for miR-30c. (C) let-7a and miR-30c nanoluciferase reporter activities in 3T3 cells. Data represent an average of three independent transfections performed as described previously (21). (D) Luciferase assay of let-7 activity in meiotically incompetent growing (GO) and fully grown (FGO) mouse oocytes injected with 10,000 or 100,000 molecules of let-7 NanoLuc luciferase reporters; reporter expression was assayed after 20 h of culture in a medium preventing resumption of meiosis. (E) Luciferase assay of let-7 activity in fully grown bovine and porcine oocytes. A total of 10,000 molecules of NanoLuc luciferase let-7a reporters were microinjected; reporter expression was assayed after 20 h of culture. (F) Luciferase assay of let-7 activity in fully grown porcine oocytes. A total of 10,000 molecules of NanoLuc luciferase let-7a reporters were microinjected; reporter expression was assayed after 40 h of culture. (G) Luciferase assay of miR-30 activity in fully grown mouse, bovine, and porcine oocytes. A total of 10,000 molecules of NanoLuc luciferase reporters carrying 1×-perfect, 4×-bulged or 4×-mutant miR-30c binding sites were microinjected; reporter expression was assayed after 20 h of culture in a medium preventing resumption of meiosis. All luciferase data are a ratio of the NanoLuc luciferase reporter activity over a co-injected control firefly luciferase reporter activity; the relative 4×-mutant reporter was set to one. All error bars represent standard deviation (SD). Asterisks indicate statistical significance (P-value) of one-tailed t-test (* < 0.05, ** < 0.01, *** < 0.001).