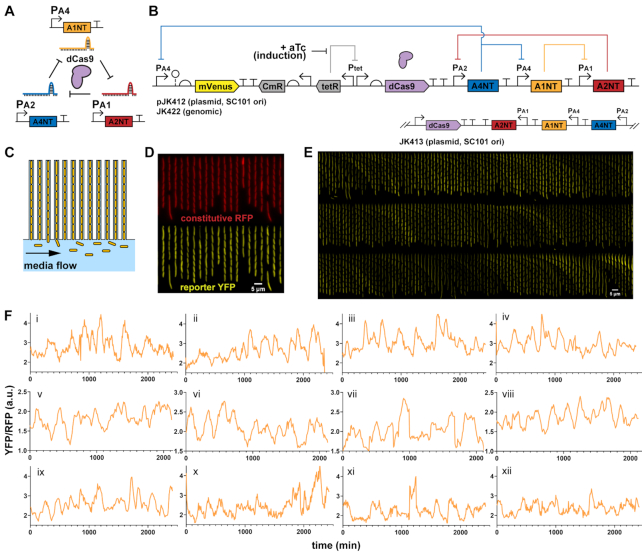
Figure 1.
dCas9 RNA oscillator imaged in microfluidic device. (A) The ring oscillator has guide RNAs targeting each other through deactivated Cas9 (dCas9), which forms a complex with a single guide RNA (sgRNA) to create an active transcriptional repressor. Each guide RNA targets the sigma factor binding region of another promoter, which produces its own sgRNA. (B) A design of the dCas9 RNA oscillator with YFP fluorescent reporter mVenus, Tet-inducible dCas9 and the sgRNA ring. The circuit is combined in a single low-copy plasmid (pSC101 origin of replication), genome-integrated or split across two plasmids. Another form has the sgRNAs transcriptionally reversed. (C) The ‘mother machine’ microfluidic device allows imaging of thousands of cells. Cells are confined in trenches, where ‘mother’ cells are perpetually trapped to allow for single-cell tracking over many generations. (D) Escherichia coli MG1655 cells with the designed dCas9 RNA oscillator plasmid (pJK412) are in a mother machine device, with constitutive RFP expressed genomically as a segmentation marker and YFP reporter expressed from the circuit. A field-of-view at one timepoint (6 h post- aTc induction) shows many cells. (E) In a kymograph looking at a single trench over time, YFP reporter fluorescence appears to fluctuate. Each frame is 8 min, with one generation ∼32 min. (F) Example representative traces of mother cell reporter YFP fluorescence normalized by segmentation RFP signal show fluctuations over time for (i–iv) 1-plasmid circuit (pJK412), (v–viii) genome-integrated circuit (JK422) and (ix–xii) 1-plasmid circuit with flipped sgRNA direction (pJK413).