Figure 4.
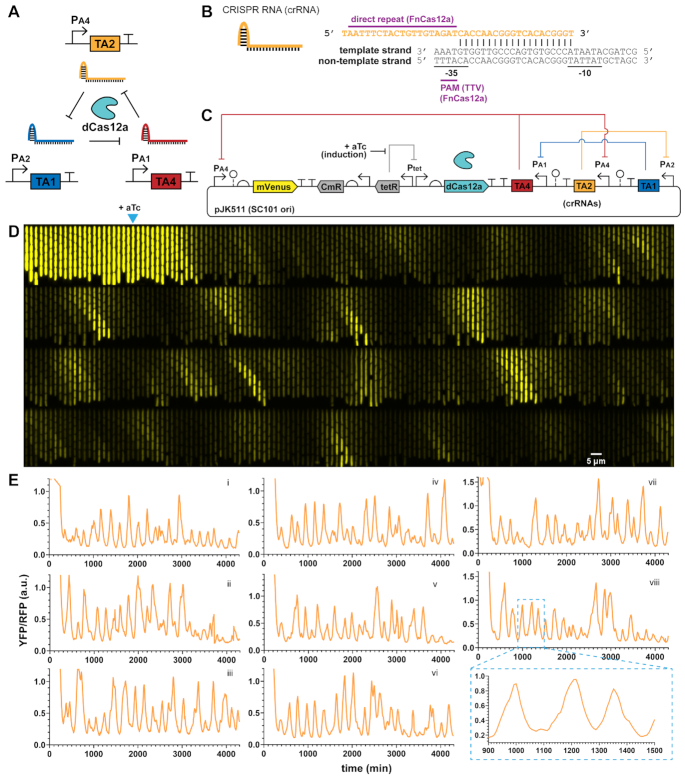
dCas12a RNA oscillator shows regularity. (A) An analogous RNA ring oscillator design replaces dCas9 with deactivated Cas12a (dCas12a). (B) Cas12a CRISPR RNA (crRNA) architecture. Designs use a 19-nt direct repeat dCas12a handle plus 20-nt targeting region. The PAM is ‘TTV’ for Francisella novicida Cas12a (FnCas12a). (C) Plasmid design of the dCas12a RNA oscillator circuit has a PA4-mVenus reporter, Tet-inducible dCas12a and crRNAs in a transcriptionally convergent direction. (D and E) Escherichia coli MG1655 cells with the circuit were grown in mother machine chips with EZ-RDM. (D) A kymograph of a single trench over time shows YFP reporter fluctuation. Each frame is 8 min, with one generation ∼25 min. Arrow marks induction for dCas12a production at frame 23 (184 min). (E) Example reporter traces show fluctuations resembling oscillations and a lower signal repressed state. Average generation ∼25 min. Pullout for trace viii zooms in on the peak shapes.