Figure 1.
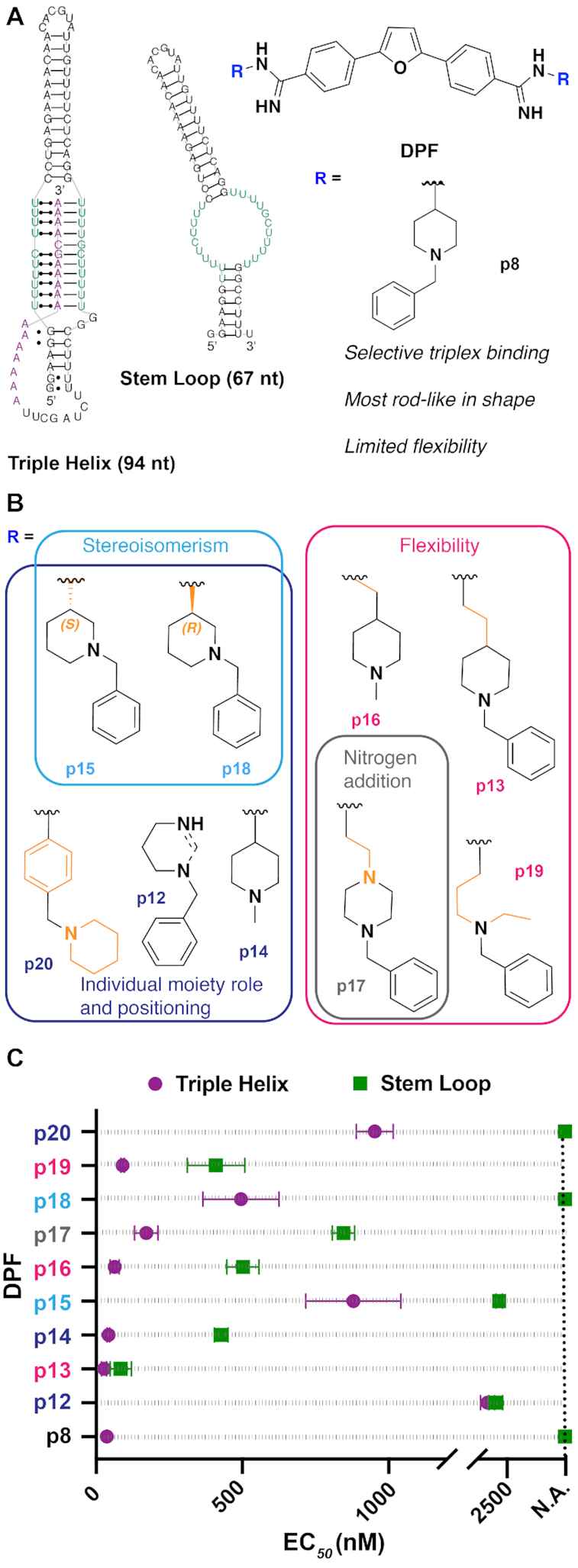
(A) Summary of the previous MALAT1-targeted study (34), in which DPF scaffold-based molecules were evaluated against the MALAT1 triple helix and stem loop structures, leading to the identification of DPFp8 as a triplex-selective ligand. nt = nucleotide. The RNA stem loop secondary structure was predicted using the default settings in the RNAstructure web server. (B) Design of the current study, in which DPFp8 analogs were designed to explore various aspects of the R group. R group number scheme reflects the expansion upon the original para-sublibrary containing subunits p1–p11. (C) EC50 values of DPFp8 analogs for binding to the MALAT1 triple helix and stem loop RNA as measured by monitoring the change in fluorescence of DPF ligands with increasing concentrations of RNA. See the ‘Materials and Methods’ section for EC50 value calculation. Values represent averages of three independent experiments ± standard deviation. Data points at dashed line indicate a not available (N.A.) value where little to no change in fluorescence of DPF (1 μM) was observed at the RNA concentrations used in the experiment (0–5 μM), leading to ambiguous curve fits.
