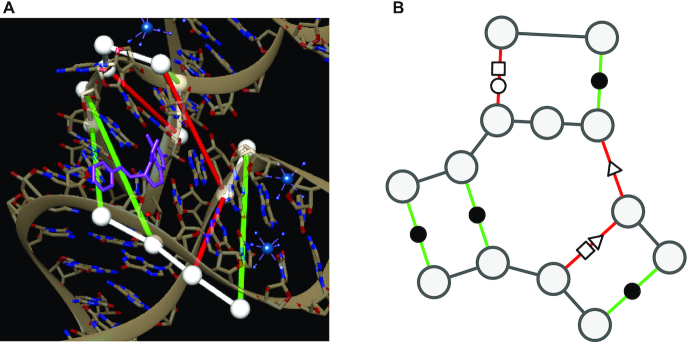
Figure 1.
(A) Binding site atomic coordinates. (B) Graph encoding of binding site as an augmented base pairing network (ABPN). RNA structure representation of the THF riboswitch binding site (PDB: 4LVV) as atomic coordinates using UCSF Chimera (23)(left) and resulting augmented base pairing network (ABPN) (right). We superpose the ABPN in the 3D visualization. Nodes are drawn as white spheres, backbone connections are in white, and canonical and non-canonical base pairs are green and red tubes respectively. We color the edges simply to guide the eye to the corresponding base pairs but note that edge color has no special meaning to our graphs. We annotate the graph representation with the standard Leontis-Westhof nomenclature for pairing type symbols. In this case, the binding site has three canonical interactions denoted (•), and three non canonicals of types (□○, ▹, □▹).