Figure 2.
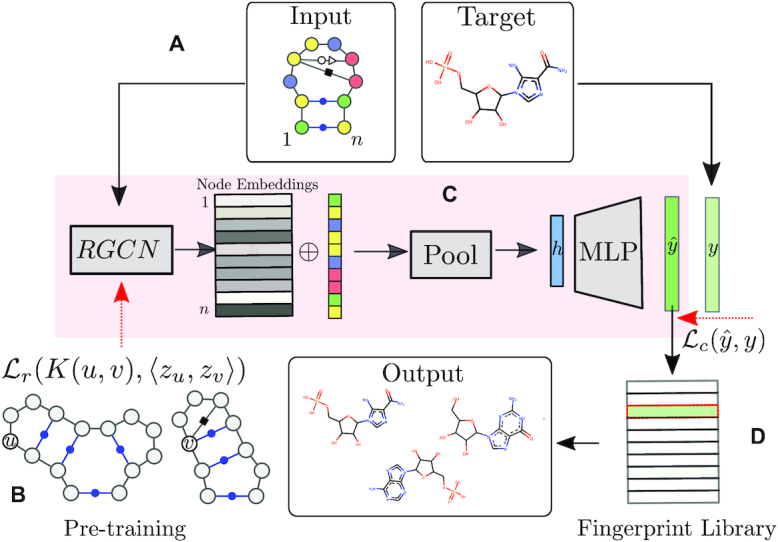
Outline of the RNAmigos pipeline. A base pairing network is passed as input to RNAmigos. In training mode, it is paired with a native ligand (Target) from which a target fingerprint y is constructed. The embedding network (RGCN) produces a matrix of node embeddings of dimension n × d where n is the number of nodes in the graph, and d is a fixed embedding size. This is followed by a pooling step which reduces node embeddings to a single graph-level vector. Finally, the graph representation is fed through a multi-layer perceptron (MLP) to produce a predicted fingerprint 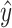 that minimizes the distance
that minimizes the distance 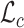 to the native fingerprint y. The fingerprint is then used to search for similar ligands to the prediction in a ligand screen and thus enriches the probability of identifying an active compound. The RGCN network is pre-trained using an unsupervised node embedding framework which allows us to leverage structural patterns from a large dataset of RNA structures. This network is trained to generate embeddings which minimize the distance (
to the native fingerprint y. The fingerprint is then used to search for similar ligands to the prediction in a ligand screen and thus enriches the probability of identifying an active compound. The RGCN network is pre-trained using an unsupervised node embedding framework which allows us to leverage structural patterns from a large dataset of RNA structures. This network is trained to generate embeddings which minimize the distance (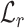 ) between kernel similarities k(u,
) between kernel similarities k(u, 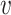 ) and embedding similarities 〈
) and embedding similarities 〈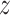 u,
u, 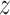
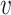 〉.
〉.
