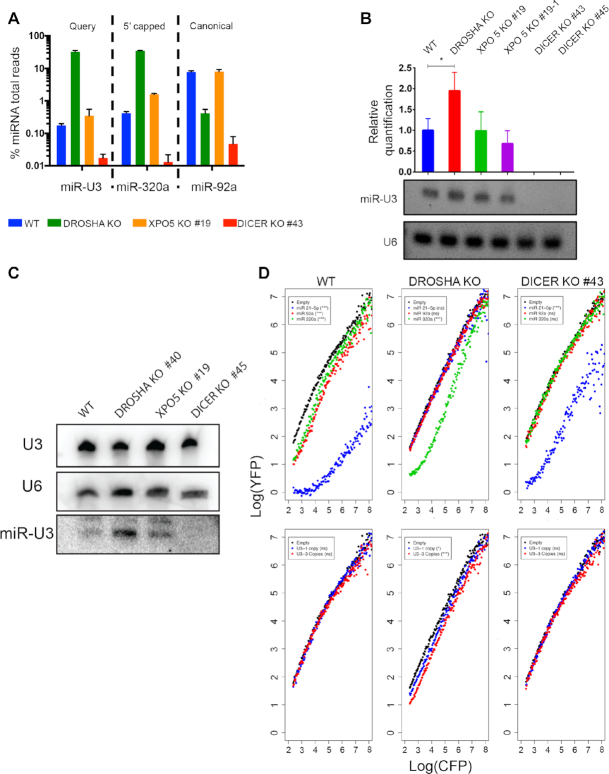
Figure 4.
Production of the U3-derived miRNA requires Dicer but not Drosha. (A) The relative proportions of sequencing reads derived from the indicated miRNAs in small RNA-seq datasets derived from wild-type HCT116 cells (WT) or HCT116 cells lacking Drosha, XPO5 or Dicer (KO) is shown. (B) Total RNA from wild-type HCT116 cells (WT) or the knockout (KO) cell lines indicated was reverse transcribed and the levels of the U3-derived miRNA and the U6 snRNA were determined using LNA-based qPCR. Data from three independent experiments is shown as mean ± standard deviation. Amplified products were separated by gel electrophoresis and detected using Ethidium bromide. (C) Total RNA was purified from wild-type HCT116 cells (WT) or HCT116 cells lacking Drosha, XPO5 or Dicer (KO) and small RNAs (<200 nt) were enriched. Total and small RNAs were separated by denaturing polyacrylamide gel electrophoresis and transferred to a nylon membrane where they were detected by northern blotting using probes against the U3 and U6 snRNAs (total RNA) and the U3-derived miRNA (small RNAs). (D) The relative expression of CFP and YFP in individual cells (dots) was determined by flow cytometry in wild-type HCT116 cells (WT) or cells lacking Drosha or Dicer (KO) containing constructs encoding no miRNA target site (Empty), or the indicated miRNA target sites in one or three copies, within the 3′ UTR of the CFP gene. Significance was calculated using the Mann–Whitney–Wilcox test; ns – not significant, *P < 0.05, **P < 0.01, *** P ≤ 0.001.