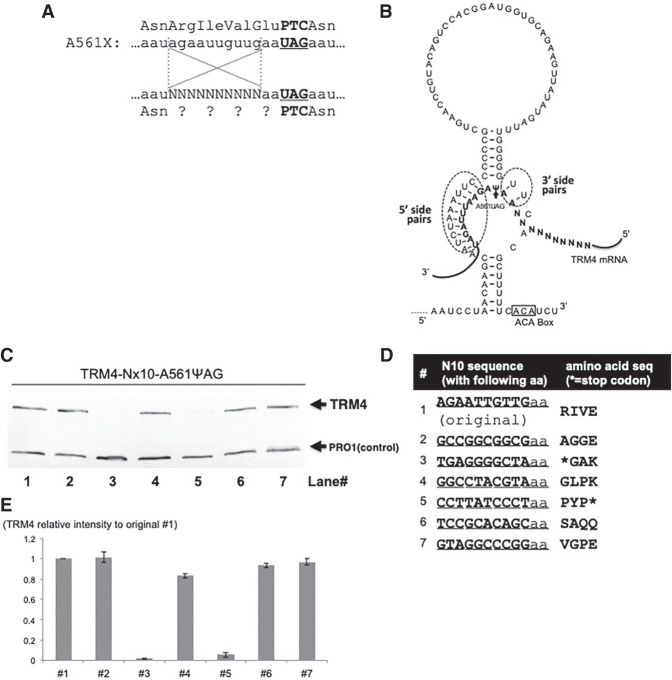
FIGURE 8.
The effect of an upstream sequence on Ψ-mediated nonsense suppression. (A) A short sequence upstream of the PTC (UAG) at codon 561 (A561X) is shown. Above the A561X nucleotide sequence is the amino acid sequence. The bold letters indicate the stop codon 561 (or PTC561). Below the A561X nucleotide sequence is a mutant A561X sequence where the 10 nt from −12 to −3 (relative to the uridine [+1] of the PTC) are randomized (Ns), and this mutant sequence was used for screen experiments. Below the mutant A561X sequence is the amino acid sequence with question marks representing unknown amino acids (due to the 10 randomized nucleotides within the A561X mRNA). (B) The 3′ hairpin of designer box H/ACA gRNA (targeting the uridine of the A561X stop codon) is shown. Also shown are the base-pairing interactions between the guide sequence and the substrate sequence (bold letters). While there are 10 bp on the 5′ side of pseudouridylation pocket (dotted circle), there are only 2 bp on the 3′ side (dotted circle). The 10 randomized nucleotides (Ns) are also shown. (C) Western blotting analysis of some representative samples (after screen experiments, see text). Yeast cells were cotransformed with the 10 nt randomized A561X construct (described in A) and the PTC-specific designer gRNA (shown in B). Several colonies were selected and total proteins were isolated from these individual colonies. Western blotting was carried out exactly as in Fig. 2A. Trm4 and Pro1 were indicated. (D) Sequences of the mutant A561X analyzed in C. The seven samples tested in C were sequenced, and their nucleotide sequences (left) as well as amino acid sequences (right) are shown. Two of the seven sequences, #3 and #5 corresponding to lanes 3 and 5 in C, have a stop codon (asterisks). (E) Quantitation of the experiments shown in C. The Trm4 signal was normalized against the signal of Pro1 (control) in each lane and compared with the readthrough signal generated from the original (wild-type) sequence (#1) (set as 1).