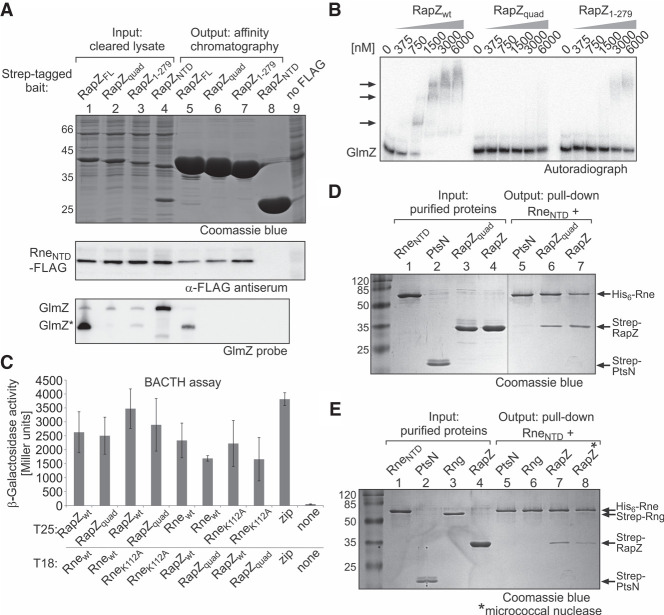
FIGURE 2.
RNA is not required for RapZ/RNase E interaction. (A) Ligand fishing experiment based on StrepTactin affinity chromatography indicating that the RNA-binding function of RapZ is dispensable for pull-down of the RneNTD. Strain Z903, which encodes a FLAG-tagged version of RneNTD in the chromosome and lacks endogenous rapZ was transformed with the following plasmids enabling overproduction of the indicated bait protein, respectively: pBGG164 (Strep-RapZFL), pYG29 (Strep-RapZquad), pSD135 (Strep-RapZ1–279), and pSD25 (Strep-RapZNTD). Cleared lysates were prepared (lanes 1–4; “input”) and subjected to StrepTactin affinity chromatography. Elution fractions (normalized to protein content; lanes 5–8) were separated on SDS-PAA gels and stained with Coomassie blue to verify successful bait protein purification (top panel) and subjected to Western analysis using anti-FLAG antiserum for detection of copurifying RneNTD (medium panel). RNA was extracted from the elution fractions and analyzed by northern blotting for presence of copurified GlmZ (bottom panel). Strain Z37 lacking the FLAG epitope served as negative control (last lane). (B) EMSA comparing the RNA-binding activities of RapZ, RapZquad, and RapZ1–279. α-32P-UTP labeled GlmZ was incubated with increasing concentrations of the indicated RapZ variant and reactions were separated on non-denaturing PAA gels and analyzed by phospho-imaging. (C) Mutations abolishing the RNA-binding activities of RapZ and RneNTD do not interfere with interaction as measured by BACTH. Tested plasmids were pBGG348 (T25-RapZ), pYG94 (T25-RapZquad), pYG101 (T25-RneNTD), pYG202 (T25-RneNTD-K112A), pYG97 (T18-RneNTD), pYG201 (T18-RneNTD-K112A), pBGG349 (T18-RapZ), and pYG39 (T18-RapZquad). Plasmid combinations pKT25-zip/pUT18C-zip and pKT25/pUT18C served as positive and negative controls, respectively. β-galactosidase activities are presented as mean ± SD. n ≥ 3. Two-tailed Student's t-tests were performed to assess whether two data sets are significantly different. The calculated P-values are reported in the source data file (Supplemental Table S3). (D) In vitro pull-down assays to probe RneNTD/RapZ interaction. Following its immobilization on Ni-NTA magnetic agarose beads, His6-RneNTD was incubated with Strep-PtsN, Strep-RapZquad, or RapZ and bound protein fractions were analyzed by SDS-PAGE/Coomassie blue staining (lanes 5–7). Purified protein preparations (“input”) were analyzed in lanes 1–4. (E) Similar approach as in (D) but with additional controls including Strep-RNase G as prey. Moreover, His6-RneNTD and Strep-RapZ preparations were treated with micrococcal nuclease prior to their coincubation (last lane). In lane 7, both proteins were treated similarly but micrococcal nuclease was omitted. Note that except for His6-RneNTD all proteins used in (D) and (E) were purified from strain Z106 (ΔglmY ΔglmZ) to exclude copurification of the sRNAs.