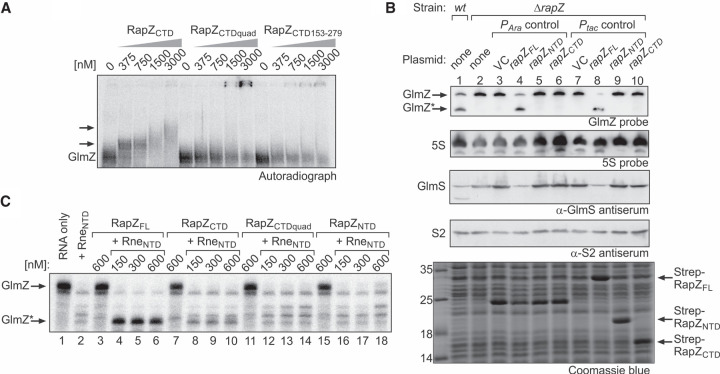
FIGURE 3.
The separated globular domains of RapZ fail to promote GlmZ cleavage in vivo and in vitro. (A) EMSA addressing the RNA-binding activity of RapZCTD and variants thereof carrying a quadruple substitution (K270A, K281A, R282A, K283A) or lacking the 5 carboxy-terminal residues. α-32P-UTP labeled GlmZ was incubated with increasing concentrations of the indicated RapZCTD variants and reactions were analyzed by native PAA gel electrophoresis followed by phospho-imaging. (B) Complementation experiment addressing the activities of plasmid-encoded RapZNTD and RapZCTD in vivo. In lanes 2–10 strain Z37 was used, which lacks endogenous rapZ, but carried plasmids triggering either low (lanes 4–6) or high (lanes 8–10) expression levels of encoded rapZ variants from the PAra or the Ptac promoter, respectively. Low level expression PAra constructs were pBAD33 (VC), pBGG61, pSD26, and pSD27. Ptac constructs triggering high expression levels were pBGG237 (VC), pBGG164, pSD25, and pSD24. In the case of the PAra constructs, gene expression was induced with 0.2% arabinose. Total RNA and protein were isolated from exponentially growing cells and subjected to northern blotting for detection of GlmZ and 5S rRNA (loading control) and western blotting for detection of GlmS and S2 (loading control) proteins. A Coomassie stained SDS-PAA gel of separated cell extracts is shown at the bottom to verify overproduction of the RapZ variants from the Ptac promoter plasmids. The wild type strain R1279 was included for comparison (lane 1). Note that the band migrating at ∼25 kDa in lanes 3–6 likely represents the chloramphenicol acetyltransferase resistance marker (MW = 24.976 kDa) encoded on pBAD33 and its derivatives. (C) In vitro RNase E cleavage assays addressing the activities of RapZCTD and RapZNTD. Radiolabeled GlmZ was coincubated with 50 nM RneNTD and increasing concentrations of RapZFL (lanes 4–6), RapZCTD (lanes 8–10), RapZCTDquad (lanes 12–14) or the RapZNTD (lanes 16–18). As negative controls, GlmZ was incubated alone (lane 1) or with each of the proteins individually (lanes 2,3,7,11,15). Following incubation, reactions were separated on denaturing PAA gels and analyzed by phospho-imaging.