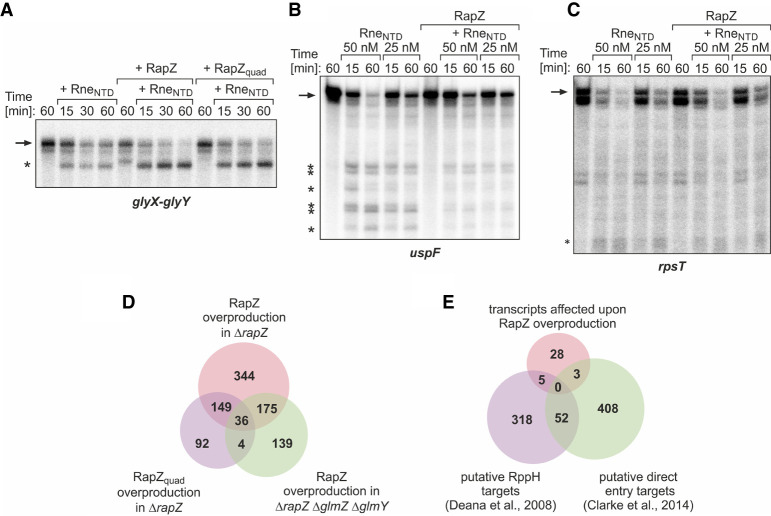
FIGURE 6.
Impact of RapZ on cleavage of non-cognate transcripts by RNase E. Time course analysis of cleavage of glyX–glyY (A), uspF (B), and rpsT (C) by RneNTD in absence and presence of RapZ in vitro. Where indicated, 600 nM RapZ or RapZquad was added to the assay. RneNTD concentrations were 5 nM for glyX–glyY cleavage assays and 50 or 25 nM for assays addressing turnover of uspF and rpsT transcripts. Samples were removed and reactions were stopped at indicated times. Reactions were separated by denaturing gel electrophoresis and analyzed by phospho-imaging. Cleavage products are indicated with asterisks. (D) Venn diagram illustrating the result of the RNA-seq analyses addressing the effect of RapZ overproduction on the transcriptome. Shown are the number of transcripts affected by overproduction of RapZ and RapZquad in strain Z37 (ΔrapZ) and of RapZ in strain Z864 (ΔrapZ ΔglmY ΔglmZ) as compared to the untransformed strains. A total of 36 transcript changes are shared by the three conditions and therefore considered to be directly caused by RapZ independent of its RNA-binding function (see also Supplemental Table S8). (E) The 36 transcripts in the intersection from (D) were compared with previous data sets reporting putative RppH targets (Deana et al. 2008) and potential direct entry RNase E substrates (Clarke et al. 2014).