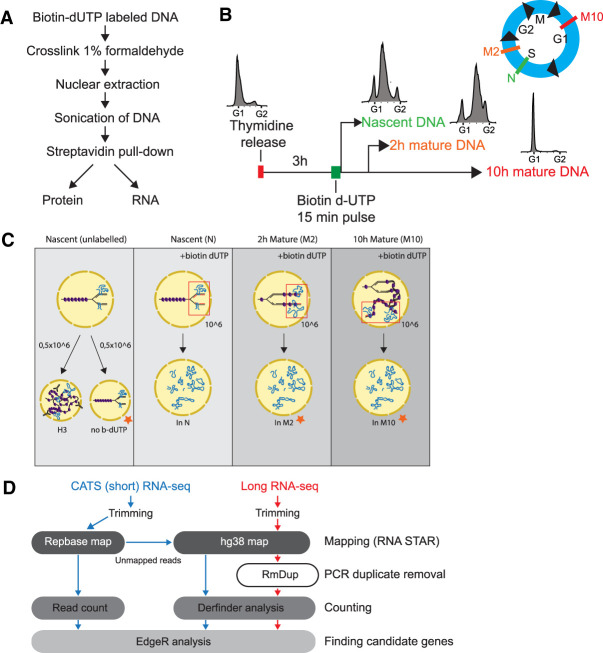
FIGURE 1.
Developing nascent chromatin capture RNA-seq (NCC RNA-seq). (A) Workflow of NCC RNA-seq protocol. (B) Experimental setup illustrating synchronization, labeling and sample harvesting. FACS diagrams showing distribution of cells in cell cycle phases at harvest time points (also indicated in blue cell cycle illustration), N = nascent chromatin, M2 = 2 h mature chromatin, M10 = 10 h mature chromatin. (C) Schematic representation of sample setup for long RNA and short RNA sequencing. Orange stars indicate samples that were included in short but not long RNA-seq. Three out of four samples were labeled 3 h into S phase with a pulse of biotin dUTP. Nascent (N) and unlabeled samples were harvested 15 min after labeling, whereas mature samples were collected 2 h (M2) and 10 h (M10) later. The unlabeled sample in short RNA-seq was split in two and used for histone 3 (H3) immunoprecipitation and as a control for unspecific binding to Streptavidin T1 beads (no b-dUTP). Input controls were taken from indicated samples before cross-linking. (D) Bioinformatics pipeline for long (red) and short (blue) NCC RNA-seq.