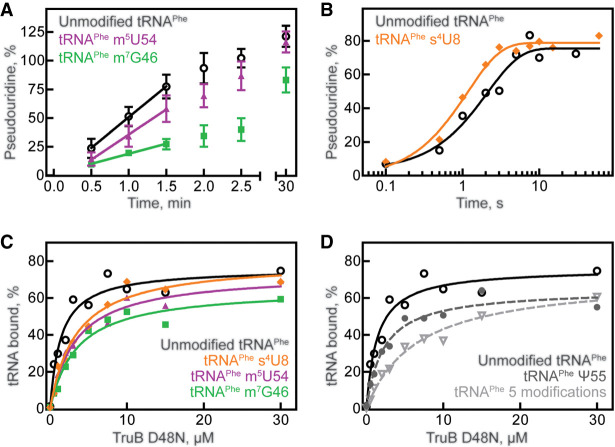
FIGURE 2.
tRNA modification and binding preferences of TruB. (A) Multiple-turnover modification by 10 nM TruB incubated with 600 nM unmodified tRNAPhe (open circles), tRNAPhe m5U54 (purple triangles), or tRNAPhe m7G46 (green squares). Time courses were performed at least in triplicate, and initial velocities were determined by linear regression (see Table 1). (B) Single-turnover modification by 5 µM TruB incubated with 1 µM unmodified tRNAPhe (open circles) or tRNAPhe s4U8 (orange diamonds). Here, we show one representative time course, but each time course was performed in triplicate and the apparent rate of each reaction was determined by fitting with a one-exponential equation, summarized in Table 2. Binding of catalytically inactive TruB D48N to 20 nM (C) unmodified tRNAPhe (open circles), tRNAPhe m5U54 (purple triangles), tRNAPhe m7G46 (green squares), tRNAPhe s4U8 (orange diamonds), or to (D) tRNAPhe Ψ55 (gray circles, dashed line), or tRNAPhe containing s4U8, Ψ32, Ψ38, m7G46, m5U54, and Ψ55 (light gray inverted triangles, dashed line), unmodified tRNAPhe (same as shown in C) determined by nitrocellulose filtration. One representative curve each of at least three replicates is shown. The data were fit with a hyperbolic equation to determine the dissociation constant, KD (see Table 3).