Figure 1.
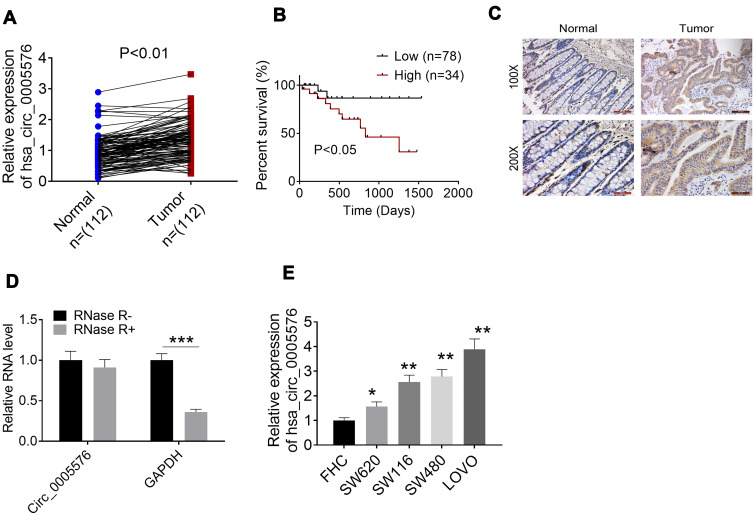
Circ_0005576 expression in CRC patients was aberrantly up-regulated. (A) qRT-PCR was used to detect circ_0005576 expression in normal/tumor tissues of 112 CRC patients. Results indicated that circ_0005576 was aberrantly up-regulated in tumor tissues compared to in normal tissues. (B) Kaplan–Meier survival analysis showed that CRC patients with high circ_0005576 expression had markedly lower percent survival than those with low circ_0005576 expression. (C) ISH assay proved the increased circ_0005576 expression in tumor tissues than that in normal tissues. (D) Circ_0005576 structural stability was researched by incubating total RNA with RNase-R. Data revealed that the circle structure of circ_0005576 could not be destroyed by RNase-R and circ_0005576 could be stably expressed in CRC. ***P < 0.001. (E) qRT-PCR illustrated that circ_0005576 expression in CRC cell lines (SW620, SW116, SW480 and LOVO) was obviously up-regulated compared to in normal colonic epithelial cell line (FHC). *P < 0.05 and **P < 0.01 when compared with FHC cells.
Abbreviations: CRC, colorectal cancer; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; qRT-PCR, quantitative real-time polymerase chain reaction; ISH, in situ hybridization.