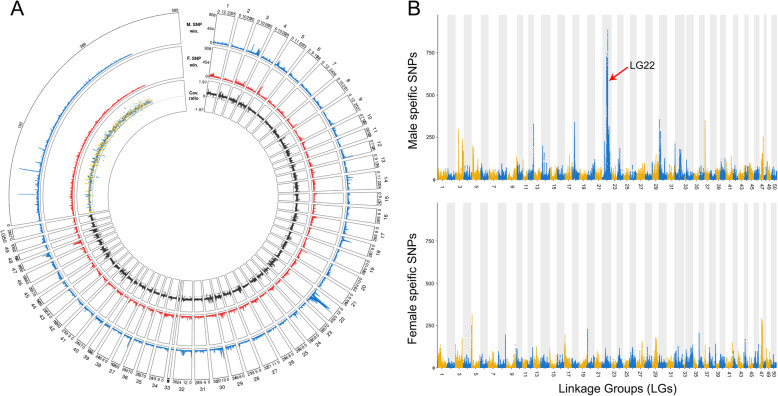
Fig. 2.
Sex determining regions identified by remapping the Pool-seq male and female reads onto the female genome assembly. SNPs were counted using 100 kb sliding window with an output point every 500 bp. a Circular plot showing the genome wide metrics of the Pool-seq analysis. All the 50 goldfish linkage groups (LGs) are labelled with their LG number and all unplaced scaffolds are fused together. Outer to inner tracks show respectively: the male-specific SNPs, the female-specific SNPs, and the reads depth ratio between males and females. b Manhattan plot of the male- and female-specific SNPs showing a strong enrichment of male-specific SNPs on LG22