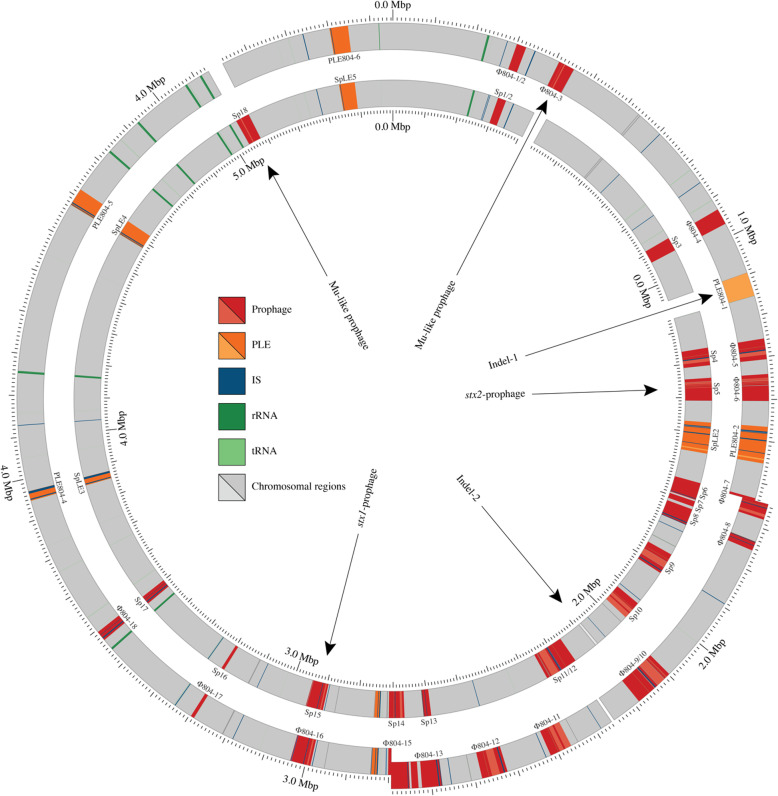
Fig. 1.
Comparison of FRIK804 and Sakai chromosomes. Alignment of the FRIK804 (outer) and Sakai (inner) chromosome found disruption of synteny by large-scale structural alterations. To evaluate each dissimilarity, the locations of relevant genomic features (prophage, PLE, IS, rRNA and tRNA) were identified. Non-conserved regions (e.g. indel-1 and indel-2) of each chromosome are shaded lighter relative to conserved regions. Evidence of mosaicism in otherwise conserved prophage was evident in several pairs of homologs, including the stx2-prophage. The majority of dissimilarities that distinguished each chromosome were associated with MGE. A 1.15 Mb inversion is denoted by the offset region in FRIK804. Indel-1 was classified as a PLE and indel-2 was not associated with any MGE. Mu-like prophage were integrated at different loci. The locations of rRNA and tRNA regions are denoted by dark green and light green regions, respectively