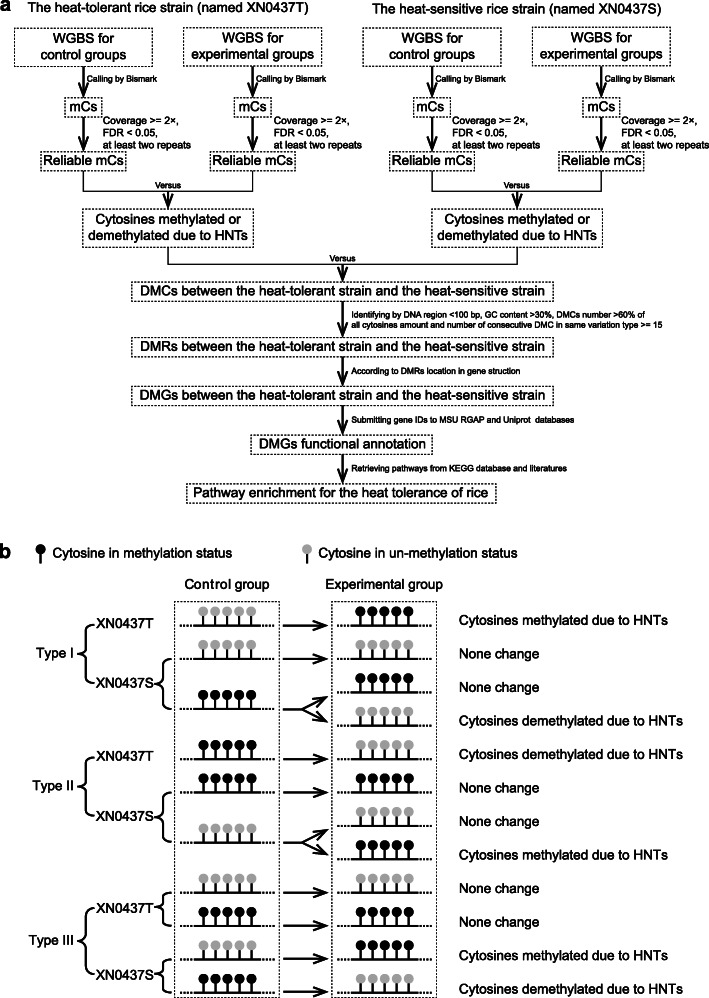
Fig. 1.
Overview of the methylation analysis workflow (a) and cytosine variation induced by high nighttime temperatures (b). WGBS, whole-genome bisulfite sequencing; HNT, high nighttime temperature; mC, methylated cytosine; DMC, a cytosine differentially methylated between the heat-tolerant and heat-sensitive coisogenic rice strains after exposure to HNTs; DMR, a DNA region < 100 bp long with a GC content of > 30%, where > 60% of all cytosines in the region were DMCs and at least 15 successive DMCs exhibited identical patterns of consistent methylation or demethylation; DMG, a differentially methylated gene adjacent to a DMR. Type I DMCs were methylated by HNTs in the heat-tolerant rice strain XN0437T, but the corresponding cytosine in the heat-sensitive rice strain XN0437S was either unchanged or demethylated after HNT. Type II DMCs were demethylated by HNTs in XN0437T, but the corresponding cytosine in XN0437S was either unchanged or methylated after HNT. Type III DMCs were unchanged by HNT in XN0437T, but the corresponding cytosine in XN0437S was either methylated or demethylated after HNT