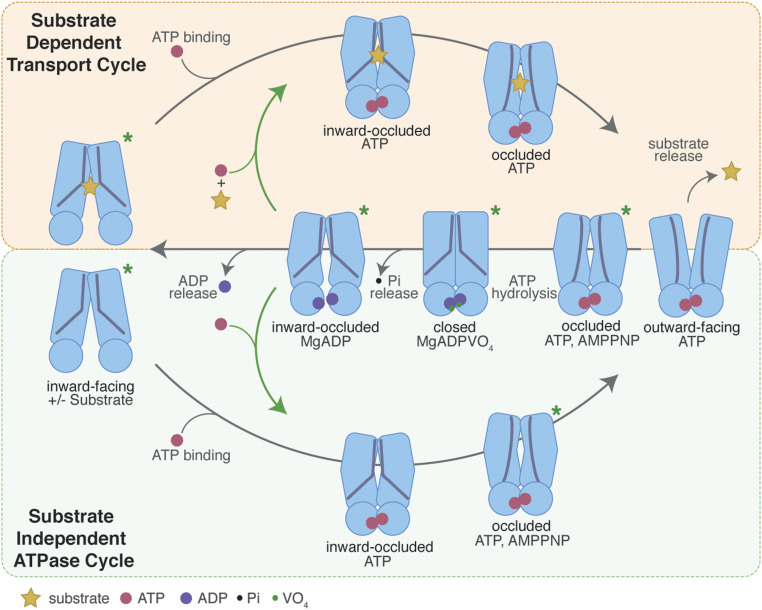
Fig. 5.
Schematic representation of a structure-based transport mechanism for NaAtm1. The substrate-dependent transport cycle (Top cycle) and the substrate-independent ATPase pathways (Bottom) differ by the presence or absence of bound substrate, respectively, during the transition from the inward- to outward-facing conformation. Resetting the outward-facing to the inward-facing conformation involves a set of intermediates common to both pathways, including the closed, posthydrolysis conformation lacking a substrate-binding site. Ligand binding to disulfide crosslinked variants may occur in the inward-occluded conformations (green arrows), without accessing the fully inward-facing conformations. Inward-facing and inward-facing occluded structures have been determined in both apo and substrate-bound states (PDB IDs: 6VQU [wide-open], 4MRN [apo], 4MRS [GSSG bound], 4MRP [GSH bound], 4MRV [GS-Hg bound], and 6PAM [MgADP bound]). The inward-occluded state with both substrate and ATP present is modeled on the crystal structure of the inward-facing occluded structure with MgADP bound (PDB ID: 6PAM). The ATP-bound occluded state has been determined for the disulfide crosslinking mutants (NaS526C and NaT525C with PDB IDs 6PAN and 6PAO, separately), the ATP hydrolysis-deficient mutant (NaE523Q with PDB ID: 6PAQ), and the wild-type transporter with MgAMPPNP bound (PDB ID: 6PAR). The outward-facing conformation of NaAtm1 has not been structurally characterized, but its presence is essential for substrate release. The posthydrolysis closed state has been characterized with MgADPVO4 bound (PDB ID: 6VQT), while the ADP-bound occluded state after ATP hydrolysis has been solved with NaA527C (PDB ID: 6PAM). Asterisks denote the structurally characterized structures to date for NaAtm1.