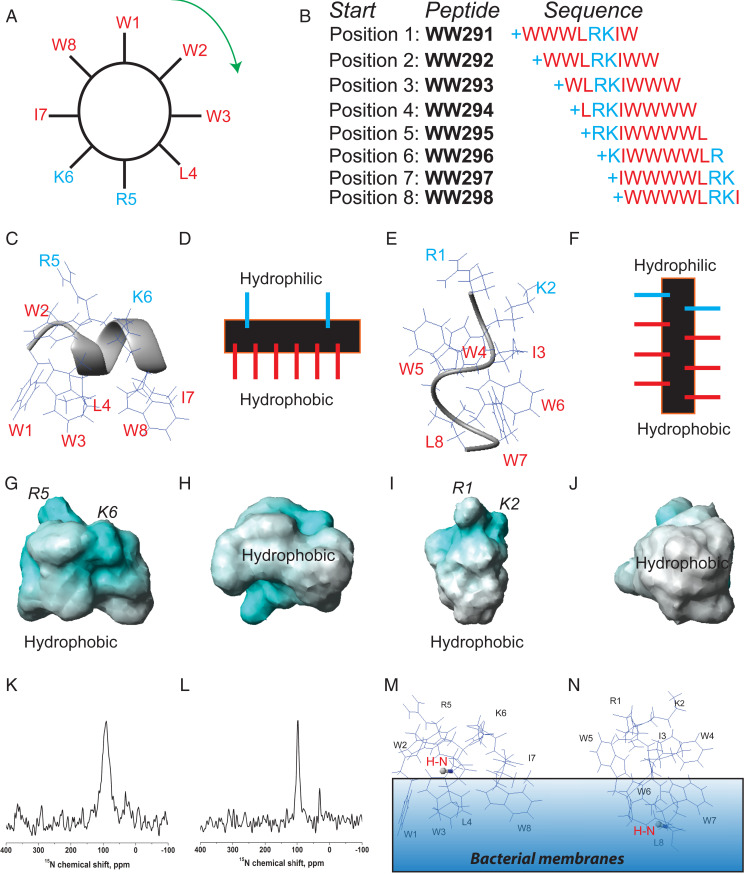
Fig. 2.
Design and NMR characterization of the two distinct amphipathic peptides. (A) The amino acid sequence of WW291 is distributed evenly on the wheel. Reading from position 1 (W1) clockwise generated WW291. Reading from position 2 (W2) in the same manner produced WW292. Repeating this reading from position 1 to position 8 (W8) led to eight peptides (B). From this sequence permutation, we identified WW295 with increased activity against Klebsiella (SI Appendix, Table S1). NMR structural determination revealed two distinct amphipathic models: Classic helix of WW291 with a horizontal axis (C and D) and nonclassic spiral structure of WW295 with a vertical axis (E and F), all relative to the hydrophobic surface. As depicted in the potential surfaces (G–J), both structures possess positive charges (cyan) for recognition of the negative bacterial surfaces and hydrophobic surfaces (white) for membrane anchoring: G and H for WW291 and I and J for WW295. NMR sample conditions are given in the legend to SI Appendix, Table S2. (K and L) Peptide membrane orientation determination by solid-state NMR spectroscopy. The solid-state NMR 15N chemical shift (<100 ppm) of a single-site 15N-labeled leucine in horine (K) and verine-L (L) indicates an H-N vector (red) parallel to membrane surface. This enables us to position the 3D structure of horine (M) and verine-L (N) on the lipid bilayer so that the H-N vector (in ball-and-stick) is approximately parallel to the bacterial membranes.