Fig. 3.
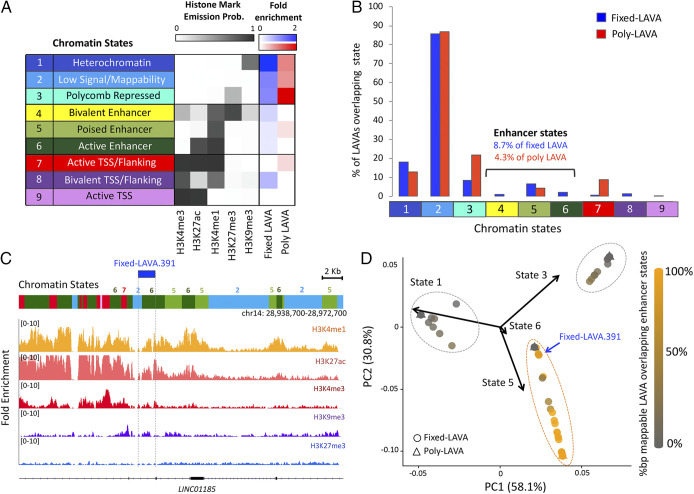
Several LAVA elements display epigenetic signatures of enhancers. (A) Chromatin states were characterized based on histone ChIP-seq signal. Fold enrichment (relative to random expectation) of states overlapping reference-present fixed (blue) and poly-LAVA (red) is shown. TSS, transcription start site. (B) Percentages of LAVA insertions with ≥10-bp overlap with each chromatin state are shown. Colors and numbers on the x axis correspond to chromatin states from A. (C) Example of a fixed LAVA that overlaps active enhancer chromatin state. ChIP-seq fold enrichment tracks (relative to input) are shown below the chromatin states. (D) PCA biplot analysis of LAVA elements based on their chromatin-state composition groups LAVA elements into three broad epigenetic groups (outlined with dashed circles). Shading elements based on their mappable percentage base pair overlap with enhancer states (state 4, 5, or 6) reveals one of these groups to be mostly composed of putatively functional LAVA (orange dashed circle). PC, principal component.