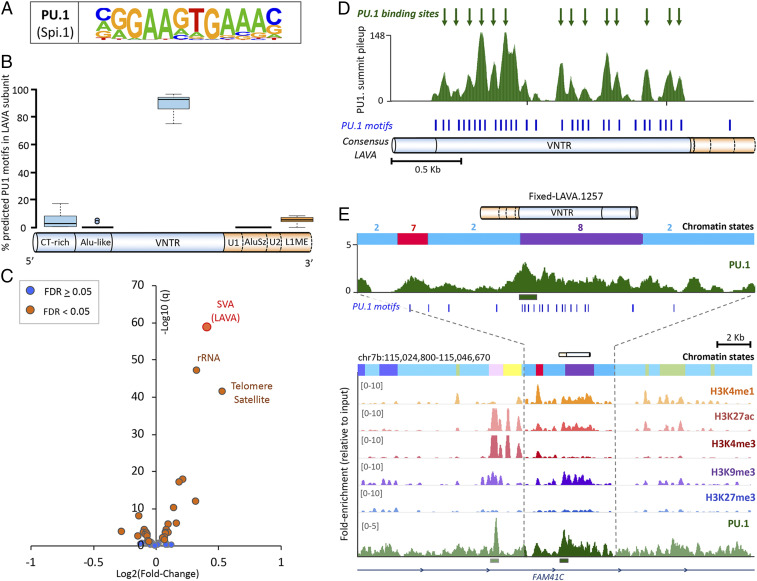
Fig. 4.
The PU.1 TF binds LAVA at the VNTR subunit. (A) PU.1 consensus binding motif from HOMER. (B) Proportions of LAVA’s predicted PU.1 binding motifs found in each subunit (based on 20 randomly selected full-length LAVA elements). U, unique nonrepetitive sequence. CT, cytosine-thymine; L1ME, long interspersed nuclear element subfamily L1ME. (C) Volcano plot displays enrichment of repeat families in gibbon PU.1 ChIP-seq. FDR, false discovery rate; rRNA, ribosomal RNA. (D) PU.1 ChIP-seq summit pileups are shown along the full-length LAVA consensus. Putative PU.1 binding sites (i.e., metasummits) are marked with green arrows. Blue ticks indicate predicted PU.1 binding motifs. (E) The fixed LAVA containing a significant PU.1 ChIP-seq peak (marked with green bar) and overlapping bivalent enhancer chromatin state (state 8 in purple). Predicted PU.1 binding motifs are marked with blue ticks. Chromatin-state colors and numbers match those in Fig. 3A.