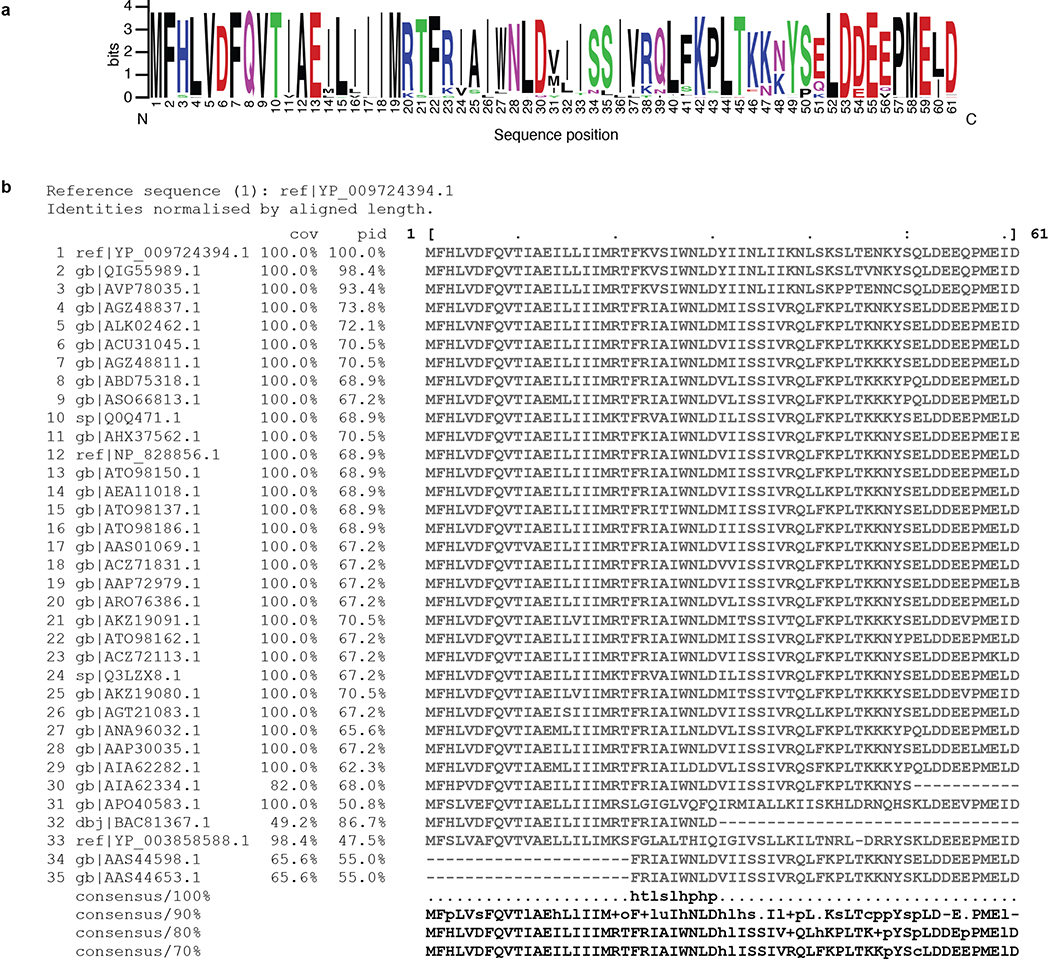
Extended Data Figure 7. Consensus analysis of SARS-CoV-2 Orf6 homologs.
(a) Sequence logo of SARS-CoV-2 Orf6 homologs, showing sequence conservation at each position computed from a multiple sequence alignment of 35 sequences. The key methionine M58, and the acidic residues E55, E59, and D61 of the putative NUP98-RAE1 binding motif are shown to be highly conserved. Homology determined from alignments to full length sequences. Colors indicated chemical properties of amino acids: polar (G, S, T, Y, C, green), neutral (Q, N, purple), basic (K, R, H, blue), acidic (D, E, red), and hydrophobic (A, V, L, I, P, W, F, M, black). (b) Multiple sequence alignment of SARS-CoV-2 Orf6 homologs. Query sequence shown at top (sequence 1 ref|YP_009724394.1). Sequence coverage (cov) and percent identity (pid) shown for each homologous sequence.