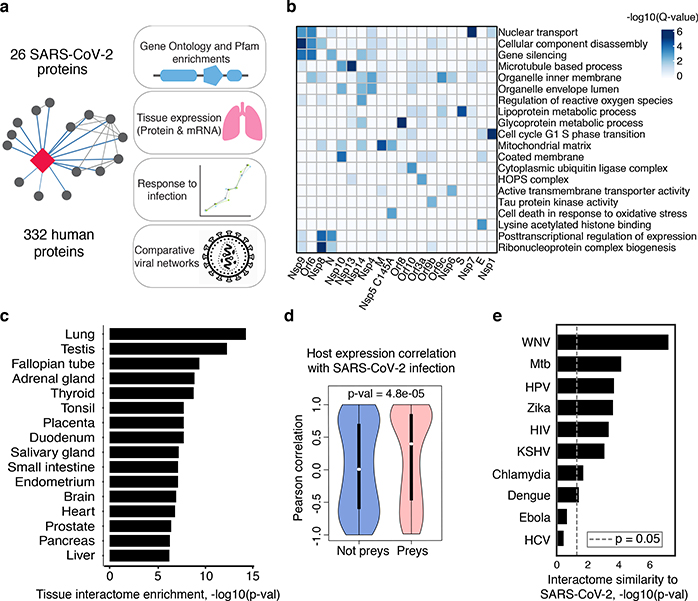
Figure 2: Global analysis of SARS-CoV-2 protein interactions.
(a) Overview of global analyses performed. (b) Gene Ontology (GO) enrichment analysis was performed on the human interacting proteins of each viral protein, p-values calculated by hypergeometric test and a false discovery rate was used to account for multiple hypothesis testing (Methods). The top GO term of each viral protein was selected for visualization. (c) Degree of differential protein expression for the human interacting proteins (n=332) across human tissues. We obtained protein abundance values for the proteome in 29 human tissues and calculated the median level of abundance for the human interacting proteins (top 16 tissues shown). This was then compared with the abundance values for the full proteome in each tissue and summarized as a Z-score from which a p-value was calculated and false discovery rate was used to account for multiple hypothesis testing. (d) The distribution of correlation of protein level changes during SARS-CoV-2 infection for pairs of viral-human proteins (median is shown) is higher than non-interacting pairs of viral-human proteins (p-value=4.8e-05, Kolmogorov–Smirnov test) The violin plots show each viral to human protein correlation for preys (n=210, min=−0.986, max=0.999, Q1=−0.468, Q2=0.396, Q3=0.850) and non-preys (n=54765, min= −0.999, max=0.999, Q1=−0.599, Q2=0.006, Q3=0.700). (e) Significance of the overlap of human interacting proteins between SARS-CoV-2 and other pathogens using a hypergeometric test (unadjusted for multiple testing). The background gene set for the test consisted of all unique proteins detected by mass spectrometry across all pathogens (n=10,181 proteins).