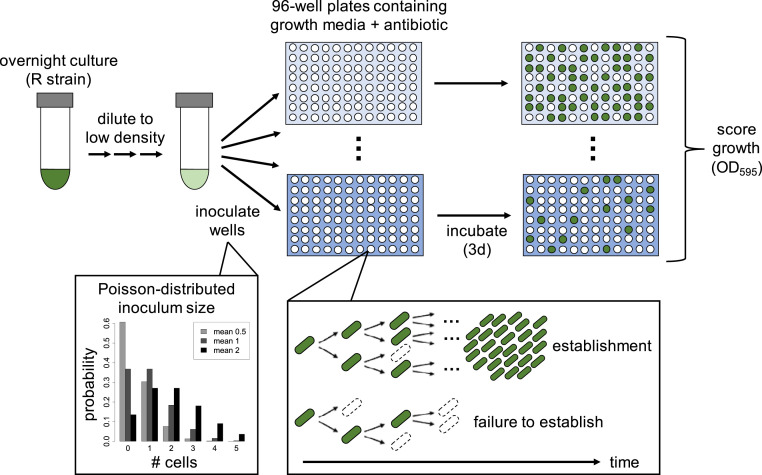
Fig. 1.
Design of seeding experiments to estimate establishment probability. An overnight culture of the resistant strain is highly diluted and used to inoculate 96-well plates containing growth media (LB broth) with antibiotic at various concentrations (shades of blue). The number of cells inoculated per well follows a Poisson distribution (examples plotted for mean inoculum size of 0.5, 1, or 2 cells per well). Within these culture wells, stochastic population dynamics imply that each inoculated cell may either produce a large number of descendants (establishment) or produce no/few descendants that ultimately die out (failure to establish). Plates are incubated for 3 d, and optical density is measured to score growth in wells (OD595 > 0.1; dark green). The number of replicate cultures showing growth is used to estimate the per-cell establishment probability at each antibiotic concentration by fitting a mathematical model.