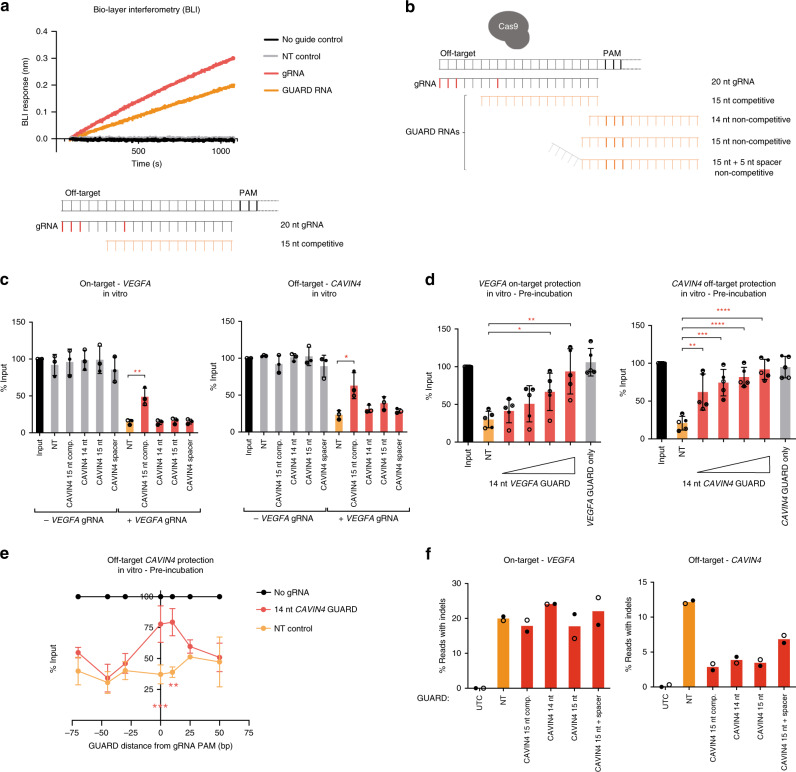
Fig. 2. GUARD RNAs reduce Cas9 off-target cleavage activity without affecting on-target editing.
a Bio-layer interferometry (BLI) analysis of binding kinetics for dead Cas9 ribonucleoprotein complex to an immobilised biotinylated DNA substrate. Cas9 was precomplexed with tracrRNA and (NT) non-targeting control gRNA; (gRNA) 20-nt VEGFA gRNA with 4 mismatches; (GUARD RNA) a 15-nt GUARD RNA targeting the off-target site or no guide RNA. b CRISPR GUARD design. Schematic showing GUARD RNAs designs. Variation in length (14-nt versus 15-nt) and protospacer positioning likely influence binding energy and competition with the on-target gRNA (i.e., competitive/overlapping, non-competitive/proximal). Mismatches in the gRNA are shown in red. RNA bases overlapping the gRNA PAM are shown in bold. c Competition between on-target gRNA and GUARD RNA can be reduced by proximal positioning. On-target (VEGFA) and off-target (CAVIN4) DNA abundance was measured by qPCR following an in vitro Cas9 DNA cleavage assay. The corresponding GUARD RNA was added at a molar ratio of 5:1 to the VEGFA on-target gRNA (100 nM to 20 nM). d GUARD RNAs are more effective at blocking Cas9-mediated DNA cutting with pre-incubation in vitro. On-target (VEGFA) and off-target (CAVIN4) DNA abundance was measured by qPCR following an in vitro Cas9 DNA cleavage assay, with a 30 min pre-incubation with GUARD RNAs and Cas9, before addition of the on-target gRNA and Cas9. Increasing molar ratios of GUARD RNA to the VEGFA on-target gRNA were used (10:10, 20:10, 50:10, 100:10 nM). e GUARD RNAs are effective in blocking Cas9-mediated DNA cleavage of off-target sites within a 10 bp window of the gRNA PAM. Synthetic DNA fragments containing the VEGFA gRNA off-target site CAVIN4 positioned progressively further away from the GUARD RNA binding site were quantified by qPCR following an in vitro Cas9 DNA cleavage assay (50:10 nM GUARD RNA to gRNA ratio). f CRISPR GUARD is effective at blocking off-target editing in cells. Indel rates from NGS of amplicons from Cas9-expressing HEK293 cells transfected with VEGFA gRNA and multiple GUARD RNA designs for protection of the CAVIN4 proximal off-target site (25:25 nM GUARD RNA to gRNA ratio). Data represent the mean of two (f) independent experiments, or the mean ± SD of three (c, e) or five (d) independent experiments with symbols representing each replicate. For a, data are representative of two independent experiments. NT non-targeting gRNA, UTC untransfected control. Unpaired, two-tailed student’s t-test. For c, *P = 0.0208, **P = 0.0083. For d, *P = 0.0164, **P = 0.0021 (VEGFA) and **P = 0.007 (CAVIN4), ***P = 0.0003, ***P < 0.0001. For e, **P = 0.0041 and ***P = 0.0001. Source data are provided as a Source Data file.