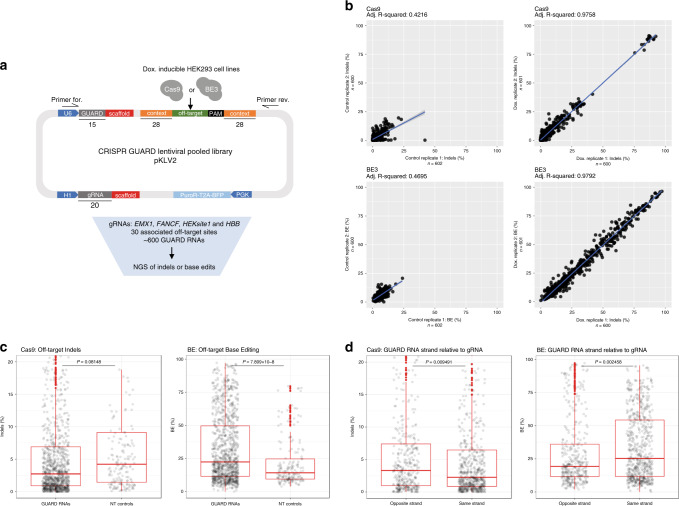
Fig. 5. High-throughput screening identifies functional GUARD RNAs.
a Schematic of the lentiviral pooled screening approach to analyse off-target editing frequencies in cells expressing different GUARD RNA species. Numbers indicate the length of the plasmid segment in base pairs. b Scatter plots of indels (Cas9) or SNPs (base edits) for two independent replicate experiments with or without induction of Cas9 or BE3 expression with doxycycline for 48 h. c Box and whiskers plot of off-target indel or base editing frequency for coding and non-targeting (NT) GUARD RNAs in the Cas9 or base editor screen, respectively. NB: some outliers fall outside of the y-axis limit. d GUARD RNAs binding to the same DNA strand as the gRNA are more effective at blocking Cas9 off-target editing but can increase base editing efficiency. Box and whiskers plot of off-target indel or SNP frequencies in the screens. NB: some outliers fall outside of the y-axis limit. Box and whiskers plot: centre line, median; box limits, upper and lower quartiles; whiskers, 1.5× interquartile range; points, outliers. Data were compared using unpaired, two-tailed student’s t-tests.