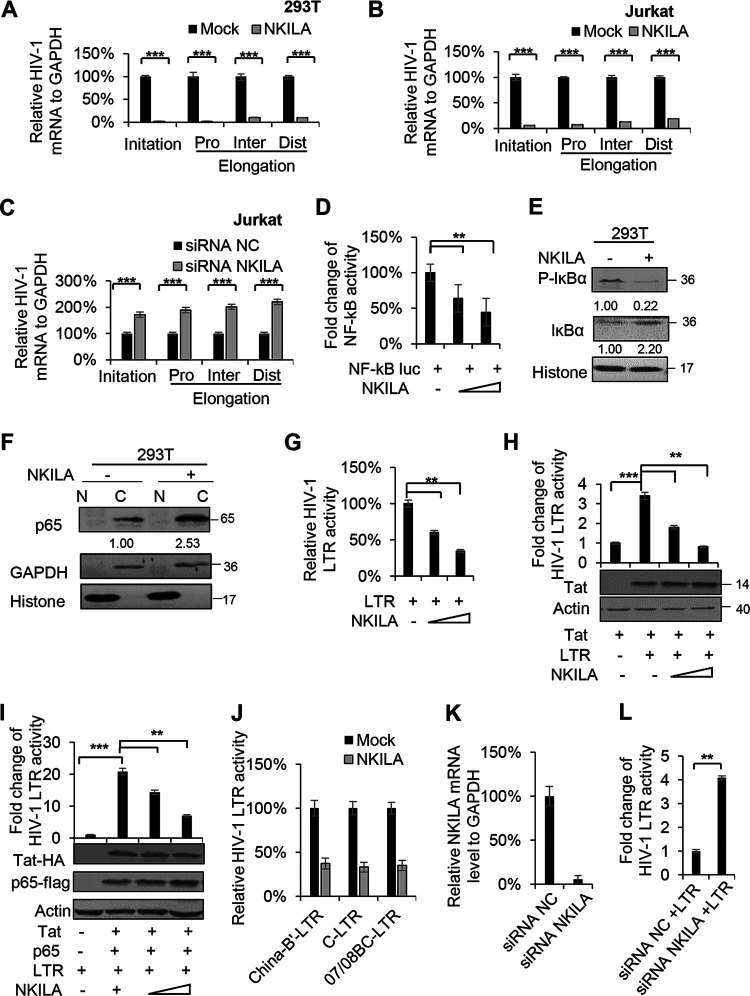
FIG 4.
NKILA suppresses HIV-1 LTR-driven gene expression in a dose-dependent manner. (A) NKILA inhibits the transcription initiation and then inhibits the transcription elongation of HIV-1 in HEK293T cells. pNL4-3 was cotransfected with NKILA or negative-control vector into HEK293T cells, and cells were harvested 48 h posttransfection for quantitation of HIV-1 gene transcription initiation and elongation with special primers by qRT-PCR. (B and C) Effect of NKILA overexpression or knockdown on HIV 5′ LTR-driven transcription initiation and elongation in Jurkat cells. Jurkat cells transfected with NKILA vectors or siRNA NKILA were infected with HIV-luc/VSV-G for 2 days, and mRNA was extracted and analyzed by qRT-PCR. (D) NKILA inhibited NF-κB activity. Increasing amounts of NKILA or negative-control vector were cotransfected with pNF-κB-luciferase and pRenilla-luciferase reporter plasmids into HEK293T cells for 48 h. Cells were harvested for assessment of reporter gene expression by a dual luciferase reporter assay. (E) NKILA expression influenced the total and phosphorylated IĸBα protein levels, as evidenced by IB analysis, and the densities of bands were analyzed with ImageJ software to calculate the values relative to that for histone. (F) The localization of p65 protein in the cytoplasm was increased with NKILA overexpression. HEK293T cells transfected with NKILA or negative-control vector were harvested for chromatin fractionation, and protein expression was then analyzed by immunoblotting. The densities of bands were analyzed with ImageJ software to calculate the values relative to that for GAPDH. (G) Overexpression of NKILA inhibited LTR-driven gene expression. Increasing amounts of NKILA or negative-control vector were cotransfected with pHIV-1-LTR-luciferase and pRenilla-luciferase into HEK293T cells for 48 h. Cells were harvested for assessment of reporter gene expression by a dual-luciferase reporter assay. NKILA also decreased LTR-driven gene expression mediated by Tat (H) or by Tat combined with p65 (I). pHIV-1 5′ LTR plasmids were cotransfected with Tat expression vector or with both Tat and p65 expression vectors. Forty-eight hours later, cell extracts were harvested for detection of protein expression by IB analysis and quantitation of reporter activity by a dual-luciferase reporter assay. (J) NKILA repressed LTR-driven gene expression of different clinical HIV-1 strains with different NF-κB binding sites. HEK293T cells were cotransfected with the LTRs of different clinical HIV-1 strains found in China and with pRenilla for 48 h. Reporter gene expression was assessed by a luciferase assay. (K and L) Knockdown of NKILA increased LTR-driven gene expression. pHIV-1 5′ LTR plasmids were cotransfected with siRNA NKILA or siRNA NC into HEK293T cells. Forty-eight hours posttransfection, the cells were harvested and analyzed by qRT-PCR to measure the mRNA level of NKILA and were subjected to a dual-luciferase reporter assay to measure the reporter activity. The corresponding value of the control was set as 1 or 100% as appropriate. All results are representative of three independent experiments. The data are presented as the means ± SDs. **, P < 0.01; ***, P < 0.001.