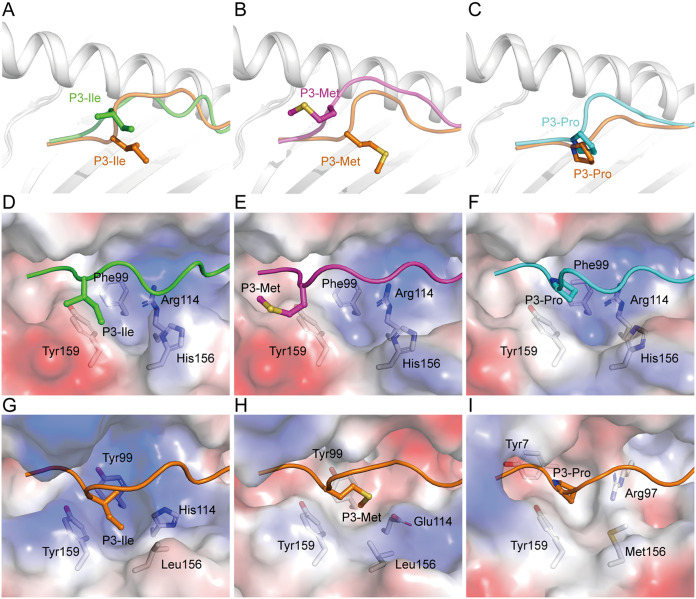
FIG 8.
The conformations of the P3 residue of peptides presented by RLA-A1. The structural comparison of the P3 residue of VP60-1 (A; green), VP60-2 (B; magenta), and VP60-10 (C; cyan) with the P3 residues of peptides in HLA-A*0201/AMPD2 (PDB code 4NO3), HLA-A*3003/MTB (PDB code 6J29), and Mamu-A*01/Tat-T18 (PDB code 1ZVS) complex, which is the same residue at the P3 position of the peptides, respectively. Peptides used for comparison are labeled in orange. The P3 residue of each peptide is shown as sticks and spheres. The backbones of MHC class I molecules are shown as cartoons in white. Vacuum electrostatic surface potentials of D pockets of RLA-A1/VP60-1 (D), RLA-A1/VP60-2 (E), RLA-A1/VP60-10 (F), HLA-A*0201/AMPD2 (G), HLA-A*3003/MTB (H), and Mamu-A*01/Tat-T18 (I) complexes are shown. Red represents negatively charged residues, blue represents positively charged residues, and gray indicates noncharged residues. Residues comprising the D pocket are shown as sticks under the vacuum electrostatic surface. The P3 residue of each peptide is shown as sticks and spheres.