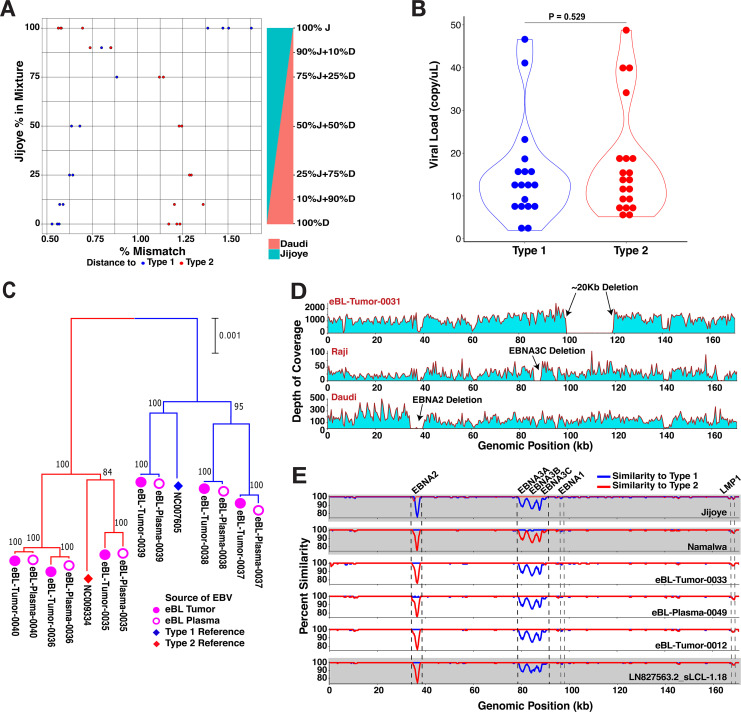
FIG 2.
Sequencing and detection quality, plasma-tumor pairs, and atypical genome isolates. (A) Controls for putative mixed infections and sampling bias against EBV types. The sensitivity of EBV genome typing approach measured by accurate type assignments of in-lab mixtures with predefined ratios. Each mixture of Daudi (type 1) and Jijoye (type 2) at various ratios was prepared in replicates. After the genome assembly, the type of the major strain was determined from the distance to both reference viral genomes. (B) Comparison of viral load levels of individuals carrying different EBV types (P = 0.529, t test). The nonsignificant difference suggests unbiased sampling among either type regardless of viral loads. We quantified viral loads with biplex qPCR using primers for viral BALF5 and human β-actin gene. (C) Comparison of virus from paired tumor (filled pink circles) and plasma samples (hollow pink circles) at diagnosis shows that viral DNA circulating in the peripheral blood represents the virus in the tumor. The neighbor-joining tree is scaled (0.001 substitutions per site) and includes standard reference genomes for type 1 (NC_007605, blue diamond) and type 2 (NC_009334, red diamond). (D) The depth of coverage showing an absence of reads from approximately 100 to 120 kb is indicative of a large deletion in the virus from an eBL tumor (top panel). In the middle and lower panels, although we did not detect any in our tumor or control viruses, we detected the deletions previously described in tumor lines, including EBNA3C deletion in Raji and EBNA2 deletion in Daudi strains. (E) Three intertypic viruses were detected by scanning across the genomes for percent identity in 1-kb windows to both type 1 and type 2 references (NC_007605 and NC_009334, respectively). The top two graphs (gray) represent the controls, Jijoye and Namalwa, followed by three intertypic viruses from this study and one publicly available intertypic virus (LN827563.2_sLCL-1.18 in gray).